- Record: found
- Abstract: found
- Article: found
Vasoactive intestinal peptide controls the suprachiasmatic circadian clock network via ERK1/2 and DUSP4 signalling
Read this article at
Abstract
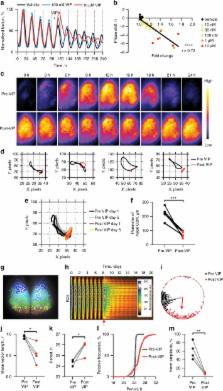
The suprachiasmatic nucleus (SCN) co-ordinates circadian behaviour and physiology in mammals. Its cell-autonomous circadian oscillations pivot around a well characterised transcriptional/translational feedback loop (TTFL), whilst the SCN circuit as a whole is synchronised to solar time by its retinorecipient cells that express and release vasoactive intestinal peptide (VIP). The cell-autonomous and circuit-level mechanisms whereby VIP synchronises the SCN are poorly understood. We show that SCN slices in organotypic culture demonstrate rapid and sustained circuit-level circadian responses to VIP that are mediated at a cell-autonomous level. This is accompanied by changes across a broad transcriptional network and by significant VIP-directed plasticity in the internal phasing of the cell-autonomous TTFL. Signalling via ERK1/2 and tuning by its negative regulator DUSP4 are critical elements of the VIP-directed circadian re-programming. In summary, we provide detailed mechanistic insight into VIP signal transduction in the SCN at the level of genes, cells and neural circuit.
Abstract
The suprachiasmatic nucleus (SCN) synchronises daily rhythms of behaviour and physiology to the light-dark cycle. Vasoactive intestinal peptide (VIP) is important for mediating SCN entrainment; however, the underlying mechanisms are incompletely understood. Here, the authors show that the effects of VIP on the SCN are mediated by ERK1/2 and DUSP4.
Related collections
Most cited references73

- Record: found
- Abstract: found
- Article: found
Sensitive red protein calcium indicators for imaging neural activity
- Record: found
- Abstract: found
- Article: not found
In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9.
- Record: found
- Abstract: found
- Article: not found