- Record: found
- Abstract: found
- Article: found
A trace fossil made by a walking crayfish or crayfish-like arthropod from the Lower Jurassic Moenave Formation of southwestern Utah, USA
Read this article at
Abstract
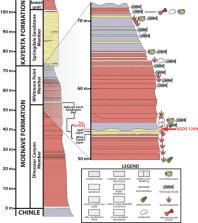
New invertebrate trace fossils from the Lower Jurassic Moenave Formation at the St. George Dinosaur Discovery Site at Johnson Farm (SGDS) continue to expand the ichnofauna at the site. A previously unstudied arthropod locomotory trace, SGDS 1290, comprises two widely spaced, thick, gently undulating paramedial impressions flanked externally by small, tapered to elongate tracks with a staggered to alternating arrangement. The specimen is not a variant of any existing ichnospecies, but bears a striking resemblance to modern, experimentally generated crayfish walking traces, suggesting a crayfish or crayfish-like maker for the fossil. Because of its uniqueness, we place it in a new ichnospecies, Siskemia eurypyge. It is the first fossil crayfish or crayfish-like locomotion trace ever recorded.
Related collections
Most cited references190
- Record: found
- Abstract: found
- Article: not found
Using Fourier transform IR spectroscopy to analyze biological materials.
- Record: found
- Abstract: not found
- Article: not found
Names for trace fossils: a uniform approach
- Record: found
- Abstract: found
- Article: not found