- Record: found
- Abstract: found
- Article: found
Expanding Imaging Capabilities for Microfluidics: Applicability of Darkfield Internal Reflection Illumination (DIRI) to Observations in Microfluidics
Read this article at
Abstract
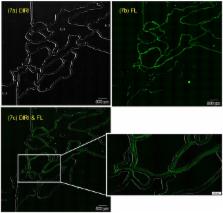
Microfluidics is used increasingly for engineering and biomedical applications due to recent advances in microfabrication technologies. Visualization of bubbles, tracer particles, and cells in a microfluidic device is important for designing a device and analyzing results. However, with conventional methods, it is difficult to observe the channel geometry and such particles simultaneously. To overcome this limitation, we developed a Darkfield Internal Reflection Illumination (DIRI) system that improved the drawbacks of a conventional darkfield illuminator. This study was performed to investigate its utility in the field of microfluidics. The results showed that the developed system could clearly visualize both microbubbles and the channel wall by utilizing brightfield and DIRI illumination simultaneously. The methodology is useful not only for static phenomena, such as clogging, but also for dynamic phenomena, such as the detection of bubbles flowing in a channel. The system was also applied to simultaneous fluorescence and DIRI imaging. Fluorescent tracer beads and channel walls were observed clearly, which may be an advantage for future microparticle image velocimetry (μPIV) analysis, especially near a wall. Two types of cell stained with different colors, and the channel wall, can be recognized using the combined confocal and DIRI system. Whole-slide imaging was also conducted successfully using this system. The tiling function significantly expands the observing area of microfluidics. The developed system will be useful for a wide variety of engineering and biomedical applications for the growing field of microfluidics.
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: not found
Droplet microfluidics.
- Record: found
- Abstract: found
- Article: not found
Detection of mutations in EGFR in circulating lung-cancer cells.
- Record: found
- Abstract: found
- Article: not found