- Record: found
- Abstract: found
- Article: found
RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability
Read this article at
Abstract
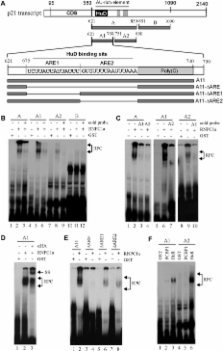
P21, a cyclin-dependent kinase inhibitor, plays a pivotal role in the cell-cycle regulation in response to stress stimuli. P21 expression is highly regulated through transcriptional, post-transcriptional and post-translational mechanisms. Previously, we and others showed that p21 expression is regulated through p21 mRNA stability by RNPC1, a target of the p53 family and HuR, a member of the ELAV family RNA-binding proteins. HuR carries three highly conserved RNA recognition motifs (RRMs) whereas RNPC1 carries one. Here we found that the ability of RNPC1 to regulate p21 mRNA stability is dependent on HuR. We also found that RNPC1 and HuR physically interact, and the RRM domain in RNPC1 and RRM3 in HuR are necessary for their interaction. Interestingly, we found that RNPC1 and HuR, both of which can bind AU-rich elements (AREs) in p21 3′-UTR, preferentially bind the upstream and downstream AREs, respectively. Finally, we showed that the RNA-binding activity of HuR to p21 transcript was enhanced by RNPC1 in vitro and in vivo. Together, we hypothesize that RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability.
Related collections
Most cited references30
- Record: found
- Abstract: not found
- Article: not found
CDK inhibitors: positive and negative regulators of G1-phase progression.
- Record: found
- Abstract: found
- Article: not found
p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities.
- Record: found
- Abstract: found
- Article: not found