- Record: found
- Abstract: found
- Article: found
Early evolution of the biotin-dependent carboxylase family
Read this article at
Abstract
Background
Biotin-dependent carboxylases are a diverse family of carboxylating enzymes widespread in the three domains of life, and thus thought to be very ancient. This family includes enzymes that carboxylate acetyl-CoA, propionyl-CoA, methylcrotonyl-CoA, geranyl-CoA, acyl-CoA, pyruvate and urea. They share a common catalytic mechanism involving a biotin carboxylase domain, which fixes a CO 2 molecule on a biotin carboxyl carrier peptide, and a carboxyl transferase domain, which transfers the CO 2 moiety to the specific substrate of each enzyme. Despite this overall similarity, biotin-dependent carboxylases from the three domains of life carrying their reaction on different substrates adopt very diverse protein domain arrangements. This has made difficult the resolution of their evolutionary history up to now.
Results
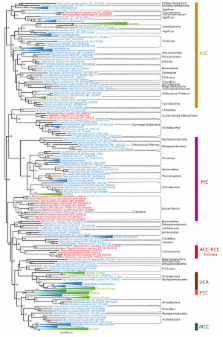
Taking advantage of the availability of a large amount of genomic data, we have carried out phylogenomic analyses to get new insights on the ancient evolution of the biotin-dependent carboxylases. This allowed us to infer the set of enzymes present in the last common ancestor of each domain of life and in the last common ancestor of all living organisms (the cenancestor). Our results suggest that the last common archaeal ancestor had two biotin-dependent carboxylases, whereas the last common bacterial ancestor had three. One of these biotin-dependent carboxylases ancestral to Bacteria most likely belonged to a large family, the CoA-bearing-substrate carboxylases, that we define here according to protein domain composition and phylogenetic analysis. Eukaryotes most likely acquired their biotin-dependent carboxylases through the mitochondrial and plastid endosymbioses as well as from other unknown bacterial donors. Finally, phylogenetic analyses support previous suggestions about the existence of an ancient bifunctional biotin-protein ligase bound to a regulatory transcription factor.
Conclusions
The most parsimonious scenario for the early evolution of the biotin-dependent carboxylases, supported by the study of protein domain composition and phylogenomic analyses, entails that the cenancestor possessed two different carboxylases able to carry out the specific carboxylation of pyruvate and the non-specific carboxylation of several CoA-bearing substrates, respectively. These enzymes may have been able to participate in very diverse metabolic pathways in the cenancestor, such as in ancestral versions of fatty acid biosynthesis, anaplerosis, gluconeogenesis and the autotrophic fixation of CO 2.
Related collections
Most cited references68
- Record: found
- Abstract: found
- Article: not found
TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics
- Record: found
- Abstract: found
- Article: not found
On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells.
- Record: found
- Abstract: not found
- Article: not found