- Record: found
- Abstract: found
- Article: found
Ubiquitin-proteasome system controls ciliogenesis at the initial step of axoneme extension
Read this article at
Abstract
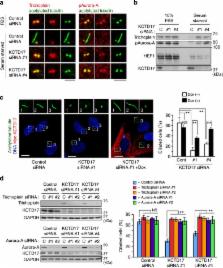
Primary cilia are microtubule-based sensory organelles that organize numerous key signals during developments and tissue homeostasis. Ciliary microtubule doublet, named axoneme, is grown directly from the distal end of mother centrioles through a multistep process upon cell cycle exit; however, the instructive signals that initiate these events are poorly understood. Here we show that ubiquitin-proteasome machinery removes trichoplein, a negative regulator of ciliogenesis, from mother centrioles and thereby causes Aurora-A inactivation, leading to ciliogenesis. Ciliogenesis is blocked if centriolar trichoplein is stabilized by treatment with proteasome inhibitors or by expression of non-ubiquitylatable trichoplein mutant (K50/57R). Started from two-stepped global E3 screening, we have identified KCTD17 as a substrate-adaptor for Cul3-RING E3 ligases (CRL3s) that polyubiquitylates trichoplein. Depletion of KCTD17 specifically arrests ciliogenesis at the initial step of axoneme extension through aberrant trichoplein-Aurora-A activity. Thus, CRL3-KCTD17 targets trichoplein to proteolysis to initiate the axoneme extension during ciliogenesis.
Abstract
 Biogenesis of the primary cilium begins after cell cycle exit, but the regulatory
steps for its formation are poorly defined. Here the authors show that proteasome-mediated
removal of the ciliogenesis inhibitor, trichoplein, from mother centrioles initiates
the first step of ciliogenesis.
Biogenesis of the primary cilium begins after cell cycle exit, but the regulatory
steps for its formation are poorly defined. Here the authors show that proteasome-mediated
removal of the ciliogenesis inhibitor, trichoplein, from mother centrioles initiates
the first step of ciliogenesis.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
Centrioles, centrosomes, and cilia in health and disease.
- Record: found
- Abstract: found
- Article: not found