- Record: found
- Abstract: found
- Article: not found
O-GlcNAc transferase invokes nucleotide sugar pyrophosphate participation in catalysis
Read this article at
Abstract
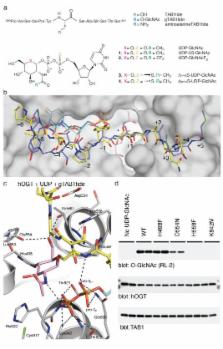
Protein O-GlcNAcylation is an essential post-translational modification on hundreds of intracellular proteins in metazoa, catalyzed by O-GlcNAc transferase using unknown mechanisms of transfer and substrate recognition. Through crystallographic snapshots and mechanism-inspired chemical probes, we define how human O-GlcNAc transferase recognizes the sugar donor and acceptor peptide and employs a novel catalytic mechanism of glycosyl transfer, involving the sugar donor α-phosphate as the catalytic base, as well as an essential lysine. This mechanism appears to be a unique evolutionary solution to the spatial constraints imposed by a bulky protein acceptor substrate, and explains the unexpected specificity of a recently reported metabolic O-GlcNAc transferase inhibitor.
Related collections
Most cited references22
- Record: found
- Abstract: found
- Article: not found
Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease.
- Record: found
- Abstract: found
- Article: not found
Structure of human O-GlcNAc transferase and its complex with a peptide substrate
- Record: found
- Abstract: found
- Article: not found
