- Record: found
- Abstract: found
- Article: found
Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment
Read this article at
Abstract
Background
Research using the zebrafish model has experienced a rapid growth in recent years. Although real-time reverse transcription PCR (QPCR), normalized to an internal reference ("housekeeping") gene, is a frequently used method for quantifying gene expression changes in zebrafish, many commonly used housekeeping genes are known to vary with experimental conditions. To identify housekeeping genes that are stably expressed under different experimental conditions, and thus suitable as normalizers for QPCR in zebrafish, the present study evaluated the expression of eight commonly used housekeeping genes as a function of stage and hormone/toxicant exposure during development, and by tissue type and sex in adult fish.
Results
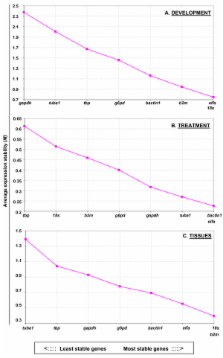
QPCR analysis was used to quantify mRNA levels of bactin1, tubulin alpha 1(tuba1), glyceraldehyde-3-phosphate dehydrogenase (gapdh), glucose-6-phosphate dehydrogenase (g6pd), TATA-box binding protein (tbp), beta-2-microglobulin (b2m), elongation factor 1 alpha (elfa) , and 18s ribosomal RNA (18s) during development (2 – 120 hr postfertilization, hpf); in different tissue types (brain, eye, liver, heart, muscle, gonads) of adult males and females; and after treatment of embryos/larvae (24 – 96 hpf) with commonly used vehicles for administration and agents that represent known environmental endocrine disruptors. All genes were found to have some degree of variability under the conditions tested here. Rank ordering of expression stability using geNorm analysis identified 18s, b2m, and elfa as most stable during development and across tissue types, while gapdh, tuba1, and tpb were the most variable. Following chemical treatment, tuba1, bactin1, and elfa were the most stably expressed whereas tbp, 18s, and b2m were the least stable. Data also revealed sex differences that are gene- and tissue-specific, and treatment effects that are gene-, vehicle- and ligand-specific. When the accuracy of QPCR analysis was tested using different reference genes to measure suppression of cyp19a1b by an estrogen receptor antagonist and induction of cyp1a by an arylhydrocarbon receptor agonist, the direction and magnitude of effects with stable and unstable genes differed.
Related collections
Most cited references37
- Record: found
- Abstract: found
- Article: not found
Housekeeping genes as internal standards: use and limits.
- Record: found
- Abstract: found
- Article: not found
Validation of housekeeping genes for normalizing RNA expression in real-time PCR.
- Record: found
- Abstract: found
- Article: not found