- Record: found
- Abstract: found
- Article: found
Rapid Diagnosis of Aneuploidy Using Segmental Duplication Quantitative Fluorescent PCR
Read this article at
Abstract
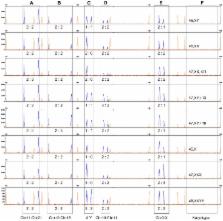
The aim of this study was use a simple and rapid procedure, called segmental duplication quantitative fluorescent polymerase chain reaction (SD-QF-PCR), for the prenatal diagnosis of fetal chromosomal aneuploidies. This method is based on the co-amplification of segmental duplications located on two different chromosomes using a single pair of fluorescent primers. The PCR products of different sizes were subsequently analyzed through capillary electrophoresis, and the aneuploidies were determined based on the relative dosage between the two chromosomes. Each primer set, containing five pairs of primers, was designed to simultaneously detect aneuploidies located on chromosomes 21, 18, 13, X and Y in a single reaction. We applied these two primer sets to DNA samples isolated from individuals with trisomy 21 (n = 36); trisomy 18 (n = 6); trisomy 13 (n = 4); 45, X (n = 5); 47, XXX (n = 3); 48, XXYY (n = 2); and unaffected controls (n = 40). We evaluated the performance of this method using the karyotyping results. A correct and unambiguous diagnosis with 100% sensitivity and 100% specificity, was achieved for clinical samples examined. Thus, the present study demonstrates that SD-QF-PCR is a robust, rapid and sensitive method for the diagnosis of common aneuploidies, and these analyses can be performed in less than 4 hours for a single sample, providing a competitive alternative for routine use.
Related collections
Most cited references19
- Record: found
- Abstract: found
- Article: not found
Segmental duplications: organization and impact within the current human genome project assembly.
- Record: found
- Abstract: found
- Article: not found
Shotgun sequence assembly and recent segmental duplications within the human genome.
- Record: found
- Abstract: not found
- Article: not found