- Record: found
- Abstract: found
- Article: not found
A Genetic Screen for Pre-mRNA Splicing Mutants of Arabidopsis thaliana Identifies Putative U1 snRNP Components RBM25 and PRP39a
Read this article at
Abstract
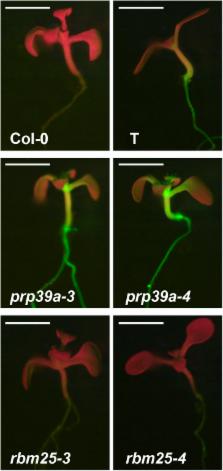
In a genetic screen for mutants showing modified splicing of an alternatively spliced GFP reporter gene in Arabidopsis thaliana, we identified mutations in genes encoding the putative U1 small nuclear ribonucleoprotein (snRNP) factors RBM25 and PRP39a. The latter has not yet been studied for its role in pre-messenger RNA (pre-mRNA) splicing in plants. Both proteins contain predicted RNA-binding domains and have been implicated in 5′ splice site selection in yeast and metazoan cells. In rbm25 mutants, splicing efficiency of GFP pre-mRNA was reduced and GFP protein levels lowered relative to wild-type plants. By contrast, prp39a mutants exhibited preferential splicing of a U2-type AT-AC intron in GFP pre-mRNA and elevated levels of GFP protein. These opposing findings indicate that impaired function of either RBM25 or PRP39a can differentially affect the same pre-mRNA substrate. Given a prior genome-wide analysis of alternative splicing in rbm25 mutants, we focused on examining the alternative splicing landscape in prp39a mutants. RNA-seq experiments performed using two independent prp39a alleles revealed hundreds of common genes undergoing changes in alternative splicing, including PRP39a itself, a second putative U1 snRNP component PRP40b, and genes encoding a number of general transcription-related proteins. The prp39a mutants displayed somewhat delayed flowering, shorter stature, and reduced seed set but no other obvious common defects under normal conditions. Mutations in PRP39b, the paralog of PRP39a, did not visibly alter GFP expression, indicating the paralogs are not functionally equivalent in this system. Our study provides new information on the contribution of PRP39a to alternative splicing and expands knowledge of plant splicing factors.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
Mechanism of alternative splicing and its regulation.
- Record: found
- Abstract: found
- Article: not found
Next-generation mapping of Arabidopsis genes.
- Record: found
- Abstract: found
- Article: not found
