- Record: found
- Abstract: found
- Article: found
Intestinal Microbiota in Patients with Spinal Cord Injury
Read this article at
Abstract
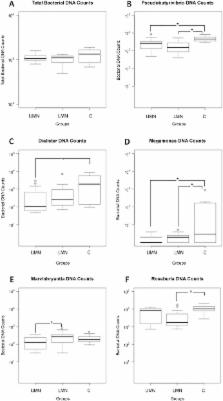
Human intestinal flora comprises thousands of bacterial species. Growth and composition of intestinal microbiota is dependent on various parameters, including immune mechanisms, dietary factors and intestinal motility. Patients with spinal cord injury (SCI) frequently display neurogenic bowel dysfunction due to the absence of central nervous system control over the gastrointestinal system. Considering the bowel dysfunction and altered colonic transit time in patients with SCI, we hypothesized the presence of a significant change in the composition of their gut microbiome. The objective of this study was to characterize the gut microbiota in adult SCI patients with different types of bowel dysfunction. We tested our hypothesis on 30 SCI patients (15 upper motor neuron [UMN] bowel syndrome, 15 lower motor neuron [LMN] bowel syndrome) and 10 healthy controls using the 16S rRNA sequencing. Gut microbial patterns were sampled from feces. Independent of study groups, gut microbiota of the participants were dominated by Blautia, Bifidobacterium, Faecalibacterium and Ruminococcus. When we compared all study groups, Roseburia, Pseudobutyrivibrio, Dialister, Marvinbryantia and Megamonas appeared as the genera that were statistically different between groups. In comparison to the healthy group, total bacterial counts of Pseudobutyrivibrio, Dialister and Megamonas genera were significantly lower in UMN bowel dysfunction group. The total bacterial count of Marvinbryantia genus was significantly lower in UMN bowel dysfunction group when compared to the LMN group. Total bacterial counts of Roseburia, Pseudobutyrivibrio and Megamonas genera were significantly lower in LMN bowel dysfunction group when compared to healthy groups. Our results demonstrate for the first time that butyrate-producing members are specifically reduced in SCI patients when compared to healthy subjects. The results of this study would be of interest since to our knowledge, microbiome-associated studies targeting SCI patients are non-existent and the results might help explain possible implications of gut microbiome in SCI.
Related collections
Most cited references18
- Record: found
- Abstract: found
- Article: not found
Towards the human intestinal microbiota phylogenetic core.
- Record: found
- Abstract: found
- Article: not found
Glial activation: a driving force for pathological pain.
- Record: found
- Abstract: found
- Article: not found