- Record: found
- Abstract: found
- Article: found
Molecular identification and subtyping of Blastocystis sp. in laboratory rats in China Translated title: Identification moléculaire et sous-typage de Blastocystis sp. chez des rats de laboratoire en Chine
Abstract
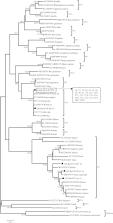
Blastocystis sp. is a ubiquitous protist that has been frequently reported in humans and animals worldwide. A total of 355 fecal samples of experimental rats were collected from four laboratory rearing facilities in China, and Blastocystis sp. was detected by PCR amplification of the partial small subunit ribosomal (SSU) rRNA gene. Twenty-nine (8.2%, 29/355) samples were positive for Blastocystis sp., with the highest infection rate (20.7%, 24/116) in rats of the Zhengzhou1, followed by that in the Zhengzhou2 (5.0%, 2/40), Shenyang (3.0%, 3/100) and Wuhan (0) rearing facilities. Among the three rat strains, Sprague–Dawley (SD) rats had higher infection rates (11.3%, 17/151) compared to Wistar rats (8.7%, 9/104) and spontaneously hypertensive (SH) rats (3.0%, 3/100). Two Blastocystis sp. subtypes (ST4 and ST7) were identified. ST4 was the predominant subtype detected in 26 samples (89.7%). A phylogenetic analysis demonstrated that the sequences of ST4 and ST7 obtained in this study were clustered with their reference subtypes. To our knowledge, this is the first report of Blastocystis sp. in experimental rats in China. Pathogen infections in laboratory animals need to be monitored due to fecal-oral transmission.
Translated abstract
Blastocystis sp. est un protiste omniprésent qui a été fréquemment signalé chez l’homme et les animaux du monde entier. Un total de 355 échantillons fécaux de rats de laboratoire ont été prélevés dans quatre installations d’élevage en laboratoire en Chine et Blastocystis sp. a été détectée par amplification par PCR du gène partiel de la petite sous-unité de l’ARNr ribosomique (SSU). Vingt-neuf échantillons (8,2 %, 29/355) étaient positifs pour Blastocystis sp., avec le taux d’infection le plus élevé (20,7 %, 24/116) chez les rats des élevages de Zhengzhou1, suivi de ceux de Zhengzhou2 (5,0 %, 2/40), Shenyang (3,0 %, 3/100) et Wuhan (0). Parmi les trois souches de rats, les rats Sprague-Dawley (SD) avaient un taux d’infection plus élevé (11,3 %, 17/151) que les rats Wistar (8,7 %, 9/104) et les rats spontanément hypertendus (SH) (3,0 %, 3/100). Deux sous-types de Blastocystis sp. ont été identifiés, ST4 et ST7. ST4 était le sous-type prédominant, détecté dans 26 échantillons (89,7 %). Une analyse phylogénétique a démontré que les séquences de ST4 et ST7 obtenues dans cette étude étaient groupées avec leurs sous-types de référence. À notre connaissance, il s’agit du premier signalement de Blastocystis sp. chez des rats de laboratoire en Chine. Les infections pathogènes chez les animaux de laboratoire doivent être surveillées en raison de la transmission fécale-orale.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype.
- Record: found
- Abstract: found
- Article: not found
Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals.

- Record: found
- Abstract: found
- Article: found