- Record: found
- Abstract: found
- Article: found
The Population Genomics of Sunflowers and Genomic Determinants of Protein Evolution Revealed by RNAseq
Read this article at
Abstract
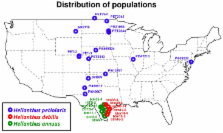
Few studies have investigated the causes of evolutionary rate variation among plant nuclear genes, especially in recently diverged species still capable of hybridizing in the wild. The recent advent of Next Generation Sequencing (NGS) permits investigation of genome wide rates of protein evolution and the role of selection in generating and maintaining divergence. Here, we use individual whole-transcriptome sequencing (RNAseq) to refine our understanding of the population genomics of wild species of sunflowers ( Helianthus spp.) and the factors that affect rates of protein evolution. We aligned 35 GB of transcriptome sequencing data and identified 433,257 polymorphic sites (SNPs) in a reference transcriptome comprising 16,312 genes. Using SNP markers, we identified strong population clustering largely corresponding to the three species analyzed here ( Helianthus annuus, H. petiolaris, H. debilis), with one distinct early generation hybrid. Then, we calculated the proportions of adaptive substitution fixed by selection ( alpha) and identified gene ontology categories with elevated values of alpha. The “response to biotic stimulus” category had the highest mean alpha across the three interspecific comparisons, implying that natural selection imposed by other organisms plays an important role in driving protein evolution in wild sunflowers. Finally, we examined the relationship between protein evolution ( d N/ d S ratio) and several genomic factors predicted to co-vary with protein evolution (gene expression level, divergence and specificity, genetic divergence [ F ST], and nucleotide diversity pi). We find that variation in rates of protein divergence was correlated with gene expression level and specificity, consistent with results from a broad range of taxa and timescales. This would in turn imply that these factors govern protein evolution both at a microevolutionary and macroevolutionary timescale. Our results contribute to a general understanding of the determinants of rates of protein evolution and the impact of selection on patterns of polymorphism and divergence.
Related collections
Most cited references50
- Record: found
- Abstract: found
- Article: not found
Adaptive protein evolution in Drosophila.
- Record: found
- Abstract: found
- Article: not found
Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees.
- Record: found
- Abstract: found
- Article: not found