- Record: found
- Abstract: found
- Article: found
Unintended Laboratory-Driven Evolution Reveals Genetic Requirements for Biofilm Formation by Desulfovibrio vulgaris Hildenborough
Read this article at
ABSTRACT
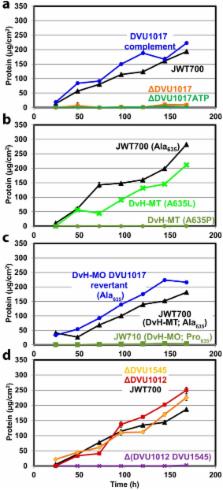
Biofilms of sulfate-reducing bacteria (SRB) are of particular interest as members of this group are culprits in corrosion of industrial metal and concrete pipelines as well as being key players in subsurface metal cycling. Yet the mechanism of biofilm formation by these bacteria has not been determined. Here we show that two supposedly identical wild-type cultures of the SRB Desulfovibrio vulgaris Hildenborough maintained in different laboratories have diverged in biofilm formation. From genome resequencing and subsequent mutant analyses, we discovered that a single nucleotide change within DVU1017, the ABC transporter of a type I secretion system (T1SS), was sufficient to eliminate biofilm formation in D. vulgaris Hildenborough. Two T1SS cargo proteins were identified as likely biofilm structural proteins, and the presence of at least one (with either being sufficient) was shown to be required for biofilm formation. Antibodies specific to these biofilm structural proteins confirmed that DVU1017, and thus the T1SS, is essential for localization of these adhesion proteins on the cell surface. We propose that DVU1017 is a member of the lapB category of microbial surface proteins because of its phenotypic similarity to the adhesin export system described for biofilm formation in the environmental pseudomonads. These findings have led to the identification of two functions required for biofilm formation in D. vulgaris Hildenborough and focus attention on the importance of monitoring laboratory-driven evolution, as phenotypes as fundamental as biofilm formation can be altered.
IMPORTANCE
The growth of bacteria attached to a surface (i.e., biofilm), specifically biofilms of sulfate-reducing bacteria, has a profound impact on the economy of developed nations due to steel and concrete corrosion in industrial pipelines and processing facilities. Furthermore, the presence of sulfate-reducing bacteria in oil wells causes oil souring from sulfide production, resulting in product loss, a health hazard to workers, and ultimately abandonment of wells. Identification of the required genes is a critical step for determining the mechanism of biofilm formation by sulfate reducers. Here, the transporter by which putative biofilm structural proteins are exported from sulfate-reducing Desulfovibrio vulgaris Hildenborough cells was discovered, and a single nucleotide change within the gene coding for this transporter was found to be sufficient to completely stop formation of biofilm.
Related collections
Most cited references36

- Record: found
- Abstract: found
- Article: found
Scalable web services for the PSIPRED Protein Analysis Workbench
- Record: found
- Abstract: found
- Article: not found
Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC.
- Record: found
- Abstract: found
- Article: not found