- Record: found
- Abstract: found
- Article: found
Development and Characterization of Polymorphic Genic-SSR Markers in Larix kaempferi
Read this article at
Abstract
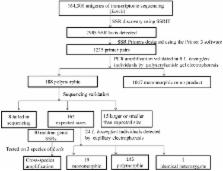
New simple sequence repeat (SSR) markers were developed in the Japanese larch ( Larix kaempferi) using unigene sequences for further genetic diversity studies and the genetic improvement of breeding programs. One thousand two handred and thirty five (1235) primer pairs were tested and 165 successfully identified in L. kaempferi. Out of the amplified candidate markers, 145 (90.6%) exhibited polymorphism among 24 individuals of L. kaempferi, with the number of alleles per locus (N a), observed heterozygosity (H o), expected heterozygosity (H e) and polymorphic information content (PIC) averaging at 4.510, 0.487, 0.518 and 0.459, respectively. Cross-species amplification of randomly selection of 30 genic-SSRs among the 145 polymorphic ones showed that 80.0% of the SSRs could be amplified in Larix olgensis, 86.7% could be amplified in Larix principi-rupprechtii and 83.0% could be amplified in Larix gmelinii. High rates of cross-species amplification were observed. The genic-SSRs developed herein would be a valuable resource for genetic analysis of Larix kaempferi and related species, and also have the potential to facilitate the genetic improvement and breeding of larch.
Related collections
Most cited references22
- Record: found
- Abstract: found
- Article: not found
Genic microsatellite markers in plants: features and applications.
- Record: found
- Abstract: not found
- Article: not found
microsatellite analyser(MSA): a platform independent analysis tool for large microsatellite data sets
- Record: found
- Abstract: found
- Article: not found