- Record: found
- Abstract: found
- Article: not found
Identification of Genes Expressed in Hyperpigmented Skin using Meta-Analysis of Microarray Datasets
Read this article at
Abstract
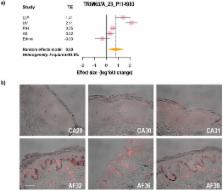
More than 375 genes have been identified that are involved in regulating skin pigmentation, and those act during development, survival, differentiation and/or responses of melanocytes to the environment. Many of those genes have been cloned and disruptions of their functions are associated with various pigmentary diseases, however many remain to be identified. We have performed a series of microarray analyses of hyperpigmented compared to less pigmented skin to identify genes responsible for those differences. The rationale and goal for this study was to perform a meta-analysis on those microarray databases to identify genes that may be significantly involved in regulating skin phenotype either directly or indirectly that might not have been identified due to subtle differences by any of those individual studies alone. The meta-analysis demonstrates that 1,271 probes representing 921 genes are differentially expressed at significant levels in the 5 microarray datasets compared, which provides new insights into the variety of genes involved in determining skin phenotype. Immunohistochemistry was used to validate 2 of those markers at the protein level (TRIM63 and QPCT) and we discuss the possible functions of those genes in regulating skin physiology.
Related collections
Most cited references20
- Record: found
- Abstract: found
- Article: not found
Physiological factors that regulate skin pigmentation.

- Record: found
- Abstract: found
- Article: found
Human pigmentation genes under environmental selection
- Record: found
- Abstract: found
- Article: not found
