- Record: found
- Abstract: found
- Article: found
Oestrogen blocks the nuclear entry of SOX9 in the developing gonad of a marsupial mammal
Read this article at
Abstract
Background
Hormones are critical for early gonadal development in nonmammalian vertebrates, and oestrogen is required for normal ovarian development. In contrast, mammals determine sex by the presence or absence of the SRY gene, and hormones are not thought to play a role in early gonadal development. Despite an XY sex-determining system in marsupial mammals, exposure to oestrogen can override SRY and induce ovarian development of XY gonads if administered early enough. Here we assess the effect of exogenous oestrogen on the molecular pathways of mammalian gonadal development.
Results
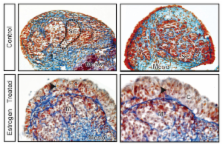
We examined the expression of key testicular ( SRY, SOX9, AMH and FGF9) and ovarian ( WNT4, RSPO1, FOXL2 and FST) markers during gonadal development in the marsupial tammar wallaby ( Macropus eugenii) and used these data to determine the effect of oestrogen exposure on gonadal fate. During normal development, we observed male specific upregulation of AMH and SOX9 as in the mouse and human testis, but this upregulation was initiated before the peak in SRY expression and 4 days before testicular cord formation. Similarly, key genes for ovarian development in mouse and human were also upregulated during ovarian differentiation in the tammar. In particular, there was early sexually dimorphic expression of FOXL2 and WNT4, suggesting that these genes are key regulators of ovarian development in all therian mammals. We next examined the effect of exogenous oestrogen on the development of the mammalian XY gonad. Despite the presence of SRY, exogenous oestrogen blocked the key male transcription factor SOX9 from entering the nuclei of male somatic cells, preventing activation of the testicular pathway and permitting upregulation of key female genes, resulting in ovarian development of the XY gonad.
Conclusions
We have uncovered a mechanism by which oestrogen can regulate gonadal development through the nucleocytoplasmic shuttling of SOX9. This may represent an underlying ancestral mechanism by which oestrogen promotes ovarian development in the gonads of nonmammalian vertebrates. Furthermore, oestrogen may retain this function in adult female mammals to maintain granulosa cell fate in the differentiated ovary by suppressing nuclear translocation of the SOX9 protein.
See commentary: http://www.biomedcentral.com/1741-7007/8/110
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: not found
Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer.
- Record: found
- Abstract: found
- Article: not found
Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation.
- Record: found
- Abstract: found
- Article: not found