- Record: found
- Abstract: found
- Article: found
Development and implementation of rapid metabolic engineering tools for chemical and fuel production in Geobacillus thermoglucosidasius NCIMB 11955
Read this article at
Abstract
Background
The thermophile Geobacillus thermoglucosidasius has considerable attraction as a chassis for the production of chemicals and fuels. It utilises a wide range of sugars and oligosaccharides typical of those derived from lignocellulose and grows at elevated temperatures. The latter improves the rate of feed conversion, reduces fermentation cooling costs and minimises the risks of contamination. Full exploitation of its potential has been hindered by a dearth of effective gene tools.
Results
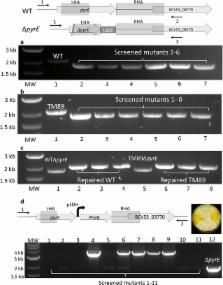
Here we designed and tested a collection of vectors (pMTL60000 series) in G. thermoglucosidasius NCIMB 11955 equivalent to the widely used clostridial pMTL80000 modular plasmid series. By combining a temperature-sensitive replicon and a heterologous pyrE gene from Geobacillus kaustophilus as a counter-selection marker, a highly effective and rapid gene knock-out/knock-in system was established. Its use required the initial creation of uracil auxotroph through deletion of pyrE using allele-coupled exchange (ACE) and selection for resistance to 5-fluoroorotic acid. The turnaround time for the construction of further mutants in this pyrE minus strain was typically 5 days. Following the creation of the desired mutant, the pyrE allele was restored to wild type, within 3 days, using ACE and selection for uracil prototrophy. Concomitant with this process, cargo DNA ( pheB) could be readily integrated at the pyrE locus. The system’s utility was demonstrated through the generation in just 30 days of three independently engineered strains equivalent to a previously constructed ethanol production strain, TM242. This involved the creation of two in-frame deletions ( ldh and pfl) and the replacement of a promoter region of a third gene ( pdh) with an up-regulated variant. In no case did the production of ethanol match that of TM242. Genome sequencing of the parental strain, TM242, and constructed mutant derivatives suggested that NCIMB 11955 is prone to the emergence of random mutations which can dramatically affect phenotype.
Conclusions
The procedures and principles developed for clostridia, based on the use of pyrE alleles and ACE, may be readily deployed in G. thermoglucosidasius. Marker-less, in-frame deletion mutants can be rapidly generated in 5 days. However, ancillary mutations frequently arise, which can influence phenotype. This observation emphasises the need for improved screening and selection procedures at each step of the engineering processes, based on the generation of multiple, independent strains and whole-genome sequencing.
Related collections
Most cited references53
- Record: found
- Abstract: found
- Article: not found
A modular system for Clostridium shuttle plasmids.
- Record: found
- Abstract: found
- Article: not found
Evolution of mutation rates in bacteria.
- Record: found
- Abstract: found
- Article: not found