- Record: found
- Abstract: found
- Article: found
Converting a Sulfenic Acid Reductase into a Disulfide Bond Isomerase
Read this article at
Abstract
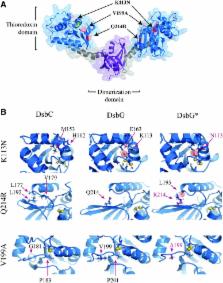
Aims: Posttranslational formation of disulfide bonds is essential for the folding of many secreted proteins. Formation of disulfide bonds in a protein with more than two cysteines is inherently fraught with error and can result in incorrect disulfide bond pairing and, consequently, misfolded protein. Protein disulfide bond isomerases, such as DsbC of Escherichia coli, can recognize mis-oxidized proteins and shuffle the disulfide bonds of the substrate protein into their native folded state. Results: We have developed a simple blue/white screen that can detect disulfide bond isomerization in vivo, using a mutant alkaline phosphatase (PhoA*) in E. coli. We utilized this screen to isolate mutants of the sulfenic acid reductase (DsbG) that allowed this protein to act as a disulfide bond isomerase. Characterization of the isolated mutants in vivo and in vitro allowed us to identify key amino acid residues responsible for oxidoreductase properties of thioredoxin-like proteins such as DsbC or DsbG. Innovation and Conclusions: Using these key residues, we also identified and characterized interesting environmental homologs of DsbG with novel properties, thus demonstrating the capacity of this screen to discover and elucidate mechanistic details of in vivo disulfide bond isomerization. Antioxid. Redox Signal. 23, 945–957.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
Protein structure alignment by incremental combinatorial extension (CE) of the optimal path.
- Record: found
- Abstract: found
- Article: not found
DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide.
- Record: found
- Abstract: found
- Article: not found