- Record: found
- Abstract: found
- Article: found
Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors
Read this article at
Abstract
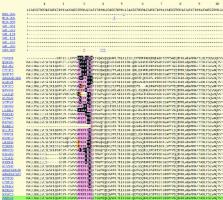
The MEROPS database ( http://merops.sanger.ac.uk) is an integrated source of information about peptidases, their substrates and inhibitors, which are of great relevance to biology, medicine and biotechnology. The hierarchical classification of the database is as follows: homologous sets of sequences are grouped into a protein species; protein species are grouped into a family; families are grouped into clans. There is a type example for each protein species (known as a ‘holotype’), family and clan, and each protein species, family and clan has its own unique identifier. Pages to show the involvement of peptidases and peptidase inhibitors in biological pathways have been created. Each page shows the peptidases and peptidase inhibitors involved in the pathway, along with the known substrate cleavages and peptidase-inhibitor interactions, and a link to the KEGG database of biological pathways. Links have also been established with the IUPHAR Guide to Pharmacology. A new service has been set up to allow the submission of identified substrate cleavages so that conservation of the cleavage site can be assessed. This should help establish whether or not a cleavage site is physiologically relevant on the basis that such a cleavage site is likely to be conserved.
Related collections
Most cited references23
- Record: found
- Abstract: found
- Article: not found
Improved tools for biological sequence comparison.

- Record: found
- Abstract: found
- Article: found
The RCSB Protein Data Bank: views of structural biology for basic and applied research and education
- Record: found
- Abstract: found
- Article: not found