- Record: found
- Abstract: found
- Article: found
Investigating the Role of Telomere and Telomerase Associated Genes and Proteins in Endometrial Cancer
Read this article at
Abstract
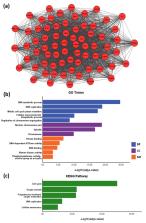
Endometrial cancer (EC) is the commonest gynaecological malignancy. Current prognostic markers are inadequate to accurately predict patient survival, necessitating novel prognostic markers, to improve treatment strategies. Telomerase has a unique role within the endometrium, whilst aberrant telomerase activity is a hallmark of many cancers. The aim of the current in silico study is to investigate the role of telomere and telomerase associated genes and proteins (TTAGPs) in EC to identify potential prognostic markers and therapeutic targets. Analysis of RNA-seq data from The Cancer Genome Atlas identified differentially expressed genes (DEGs) in EC (568 TTAGPs out of 3467) and ascertained DEGs associated with histological subtypes, higher grade endometrioid tumours and late stage EC. Functional analysis demonstrated that DEGs were predominantly involved in cell cycle regulation, while the survival analysis identified 69 DEGs associated with prognosis. The protein-protein interaction network constructed facilitated the identification of hub genes, enriched transcription factor binding sites and drugs that may target the network. Thus, our in silico methods distinguished many critical genes associated with telomere maintenance that were previously unknown to contribute to EC carcinogenesis and prognosis, including NOP56, WFS1, ANAPC4 and TUBB4A. Probing the prognostic and therapeutic utility of these novel TTAGP markers will form an exciting basis for future research.
Related collections
Most cited references234
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: found
- Article: found
Hallmarks of Cancer: The Next Generation
- Record: found
- Abstract: found
- Article: not found