- Record: found
- Abstract: found
- Article: not found
H++: a server for estimating p K as and adding missing hydrogens to macromolecules
Read this article at
Abstract
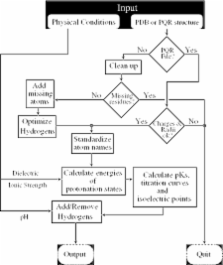
The structure and function of macromolecules depend critically on the ionization (protonation) states of their acidic and basic groups. A number of existing practical methods predict protonation equilibrium p K constants of macromolecules based upon their atomic resolution Protein Data Bank (PDB) structures; the calculations are often performed within the framework of the continuum electrostatics model. Unfortunately, these methodologies are complex, involve multiple steps and require considerable investment of effort. Our web server http://biophysics.cs.vt.edu/H++ provides access to a tool that automates this process, allowing both experts and novices to quickly obtain estimates of p Ks as well as other related characteristics of biomolecules such as isoelectric points, titration curves and energies of protonation microstates. Protons are added to the input structure according to the calculated ionization states of its titratable groups at the user-specified pH; the output is in the PQR (PDB + charges + radii) format. In addition, corresponding coordinate and topology files are generated in the format supported by the molecular modeling package AMBER. The server is intended for a broad community of biochemists, molecular modelers, structural biologists and drug designers; it can also be used as an educational tool in biochemistry courses.
Related collections
Most cited references39
- Record: found
- Abstract: found
- Article: not found
Classical electrostatics in biology and chemistry.
- Record: found
- Abstract: found
- Article: not found
Electrostatic aspects of protein-protein interactions.
- Record: found
- Abstract: found
- Article: not found
