- Record: found
- Abstract: found
- Article: found
The Importance of Biologic Knowledge and Gene Expression Context for Genomic Data Interpretation

Read this article at
Abstract
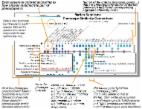
Background: Genomic sequencing, including whole exome sequencing (WES), is enabling a higher resolution for defining diseases, understand mechanisms, and improving the practice of clinical care. However, WES routinely identifies genomic variants with uncertain functional effects. Furthering uncertainty in WES data interpretation is that many genes can express multiple transcripts and their relative expression may differ by body tissue. In order to interpret WES data, we not only need to understand which transcript is most relevant, but what tissue is most relevant.
Methods: In this work, we quantify how frequently differences in transcript and tissue expression affect WES data interpretation at gene, pathway, disease, and biologic network levels. We combined and analyzed multiple large and publically available datasets to inform genomic data interpretation.
Results: Across well-established biologic pathways and genes with pathogenic disease variants, 54 and 40% have a different protein coding effect by transcript selection for, respectively, 25 and 50% of the genes contained. Additionally, strong differences in human tissue expression levels affect 33 and 19% of the same set of pathways and diseases for, respectively, 25 and 50% of the genes contained.
Conclusion: Whole exome sequencing identifies genomic variants, but to interpret the functional effects of those variants in high-resolution, we recommend building transcript selection and cross-tissue gene expression levels into hypotheses and analyses. Using current large-scale data, we show how extensively interpretation of genomic variants may differ according to transcript and tissue, across most pathways and disease. Thus, their inclusion is necessary for WES data interpretation.
Related collections
Most cited references20
- Record: found
- Abstract: found
- Article: found
The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species
- Record: found
- Abstract: found
- Article: not found
Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation.

- Record: found
- Abstract: found
- Article: found