- Record: found
- Abstract: found
- Article: found
MicroRNAs Expression in Triple Negative vs Non Triple Negative Breast Cancer in Tunisia: Interaction with Clinical Outcome
Read this article at
Abstract
Introduction
MicroRNAs are small, non coding regulatory molecules containing approximately 21 to 25 nucleotides. They function as controllers of expression at post transcriptional levels of most human protein-coding genes and play an essential role in cell signaling pathways. The objective of the present study is to evaluate the expression profile of the following micro-RNAs: miR-10b, miR-17, miR-21, miR-34a, miR-146a, miR-148a and miR-182, and to determine their possible interaction in triple-negative and non triple-negative primary breast cancers based on clinical outcome.
Methods
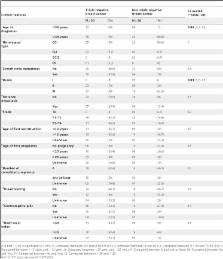
60 triple-negative and non triple-negative breast cancer cases, along with their corresponding normal samples were investigated in relation to the expression of the seven studied miRNAs using qPCR Syber Green.
Results
We observed that miR-21, miR-146a and miR-182 were significantly over expressed in triple negative breast cancer. Moreover, miR-10b, miR-21 and miR-182 were significantly associated to lymph node metastases occurrence in triple negative breast carcinoma while only miR-10b was associated with grade III in non triple negative breast cancer cases. Almost all the analyzed microRNAs were strongly associated with patients’ genico-obstetric history in non triple negative breast cancer cases except for miR-34a. All the studied microRNAs were strongly correlated with the use of the contraceptive pills in non triple negative breast cancer groups. The additive effect of hormonal factors in triple negative breast cancer cases showed an association with all the studied miRs except for miR-34 and miR-146a.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC.
- Record: found
- Abstract: found
- Article: not found