- Record: found
- Abstract: not found
- Article: not found
Efficient replacement of plasma membrane outer leaflet phospholipids and sphingolipids in cells with exogenous lipids
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
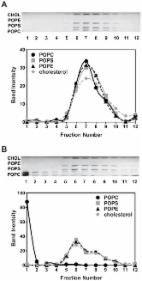
<p id="d1727670e178">The use of methyl-α-cyclodextrin to fully replace endogenous
sphingolipids and phospholipids
in plasma membrane outer leaflets of living mammalian cells with exogenous natural
and unnatural lipids is introduced in this paper. Outer leaflet lipid species were
evaluated, with lipids exchanged out of the plasma membrane outer leaflet matching
expected lipid asymmetry. The method will make possible a range of membrane structure/function
studies.
</p><p class="first" id="d1727670e181">Our understanding of membranes and membrane
lipid function has lagged far behind that
of nucleic acids and proteins, largely because it is difficult to manipulate cellular
membrane lipid composition. To help solve this problem, we show that methyl-α-cyclodextrin
(MαCD)-catalyzed lipid exchange can be used to maximally replace the sphingolipids
and phospholipids in the outer leaflet of the plasma membrane of living mammalian
cells with exogenous lipids, including unnatural lipids. In addition, lipid exchange
experiments revealed that 70–80% of cell sphingomyelin resided in the plasma membrane
outer leaflet; the asymmetry of metabolically active cells was similar to that previously
defined for erythrocytes, as judged by outer leaflet lipid composition; and plasma
membrane outer leaflet phosphatidylcholine had a significantly lower level of unsaturation
than phosphatidylcholine in the remainder of the cell. The data also provided a rough
estimate for the total cellular lipids residing in the plasma membrane (about half).
In addition to such lipidomics applications, the exchange method should have wide
potential for investigations of lipid function and modification of cellular behavior
by modification of lipids.
</p>
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: found
Preparation of Artificial Plasma Membrane Mimicking Vesicles with Lipid Asymmetry
Qingqing Lin, Erwin London (2014)
- Record: found
- Abstract: found
- Article: found
Editorial
Mario González-Quevedo, Nancy Chabot, Jasmine Thum … (1999)
- Record: found
- Abstract: found
- Article: not found
Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies.
Raphael Zidovetzki, Irena Levitan (2007)
