- Record: found
- Abstract: found
- Article: found
Prunus genetics and applications after de novo genome sequencing: achievements and prospects
Read this article at
Abstract
Prior to the availability of whole-genome sequences, our understanding of the structural and functional aspects of Prunus tree genomes was limited mostly to molecular genetic mapping of important traits and development of EST resources. With public release of the peach genome and others that followed, significant advances in our knowledge of Prunus genomes and the genetic underpinnings of important traits ensued. In this review, we highlight key achievements in Prunus genetics and breeding driven by the availability of these whole-genome sequences. Within the structural and evolutionary contexts, we summarize: (1) the current status of Prunus whole-genome sequences; (2) preliminary and ongoing work on the sequence structure and diversity of the genomes; (3) the analyses of Prunus genome evolution driven by natural and man-made selection; and (4) provide insight into haploblocking genomes as a means to define genome-scale patterns of evolution that can be leveraged for trait selection in pedigree-based Prunus tree breeding programs worldwide. Functionally, we summarize recent and ongoing work that leverages whole-genome sequences to identify and characterize genes controlling 22 agronomically important Prunus traits. These include phenology, fruit quality, allergens, disease resistance, tree architecture, and self-incompatibility. Translationally, we explore the application of sequence-based marker-assisted breeding technologies and other sequence-guided biotechnological approaches for Prunus crop improvement. Finally, we present the current status of publically available Prunus genomics and genetics data housed mainly in the Genome Database for Rosaceae (GDR) and its updated functionalities for future bioinformatics-based Prunus genetics and genomics inquiry.
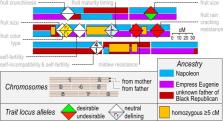
A fruitful genomic resource
Detailed genomic maps from the fruit-bearing trees of the Prunus genus should greatly accelerate the breeding of superior cultivars. These plants produce a variety of popular fruits, but until relatively recently, breeders had only limited genetic information with which to work. Pere Arús of Spain’s Institute of Agrifood Research and Technology and colleagues have now reviewed the rapid advances in Prunus genomics that have taken place since 2010, when researchers completed the first genome assembly for this genus. In the ensuing decade, increasingly high-quality genomes from the peach, Japanese apricot, and cherry have allowed scientists to home in on chromosomal regions associated with key traits such as fruit quality and disease resistance. These data also offer valuable genetic markers that agronomists can use to guide the production of healthier trees and more commercially desirable fruits.
Related collections
Most cited references159
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
fw2.2: a quantitative trait locus key to the evolution of tomato fruit size.
- Record: found
- Abstract: found
- Article: not found