- Record: found
- Abstract: found
- Article: found
RNA Sequence Analysis of Human Huntington Disease Brain Reveals an Extensive Increase in Inflammatory and Developmental Gene Expression
Read this article at
Abstract
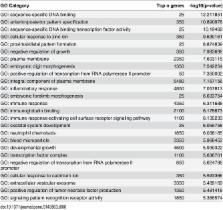
Huntington’s Disease (HD) is a devastating neurodegenerative disorder that is caused by an expanded CAG trinucleotide repeat in the Huntingtin ( HTT) gene. Transcriptional dysregulation in the human HD brain has been documented but is incompletely understood. Here we present a genome-wide analysis of mRNA expression in human prefrontal cortex from 20 HD and 49 neuropathologically normal controls using next generation high-throughput sequencing. Surprisingly, 19% (5,480) of the 28,087 confidently detected genes are differentially expressed (FDR<0.05) and are predominantly up-regulated. A novel hypothesis-free geneset enrichment method that dissects large gene lists into functionally and transcriptionally related groups discovers that the differentially expressed genes are enriched for immune response, neuroinflammation, and developmental genes. Markers for all major brain cell types are observed, suggesting that HD invokes a systemic response in the brain area studied. Unexpectedly, the most strongly differentially expressed genes are a homeotic gene set (represented by Hox and other homeobox genes), that are almost exclusively expressed in HD, a profile not widely implicated in HD pathogenesis. The significance of transcriptional changes of developmental processes in the HD brain is poorly understood and warrants further investigation. The role of inflammation and the significance of non-neuronal involvement in HD pathogenesis suggest anti-inflammatory therapeutics may offer important opportunities in treating HD.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
The calpain system.
- Record: found
- Abstract: found
- Article: not found