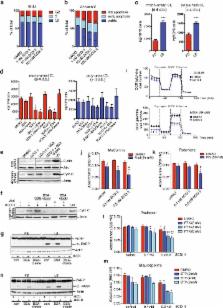
Significance Tumors are situated in a metabolically challenging environment where blood supply, and the supply of oxygen and other nutrients that come with it, is scarce. We found that nearly 40% of invasive ductal carcinoma have high expression of acetyl-CoA synthetase 2 (ACSS2). ACSS2, an enzyme that converts acetate to acetyl-CoA, imparts a competitive growth advantage under conditions of metabolic stress by enhancing the ability of cancer cells to use acetate as an additional nutritional source when other carbon sources are scarce or cannot be used to sustain lipid biomass production. This study also identified ACSS2 as a potential molecular target for managing cancer growth. Introduction Under typical cell culture conditions and in many tissues within the human body, the primary source of lipids for membrane biogenesis is the plasma. However, exposure to lipid-deplete conditions causes a highly coordinated reorganization of the lipid metabolism machinery that is primarily orchestrated by the sterol regulatory element-binding protein (SREBF) family of transcription factors, which drive the expression of genes encoding for enzymes involved in fatty acid and cholesterol biosynthesis. Deregulated expression of many of these metabolic enzymes is a feature of different disease states, including cancer. Importantly, de novo lipogenesis is a key component of anabolic metabolism and is necessary to meet the demands for biomass production required for growth and survival under unfavorable conditions (Baenke et al., 2013). Despite knowledge of the relationship between tumor progression and changes in lipid metabolism (Menendez and Lupu, 2007), it was not until the early 1990s that fatty acid synthase (FASN) was strongly associated with recurrence, metastases, and death in breast cancer patients (Kuhajda et al., 1994). Subsequently, de novo fatty acid synthesis was found to be a critical regulator of breast, prostate, and lung cancer growth (Alli et al., 2005; Orita et al., 2008; Pizer et al., 1996a, 1996b, 1996c, 2001; Puig et al., 2009; Zhan et al., 2008). Selective activation of the fatty-acid-synthesis pathway commonly occurs in many cancer types, and, in particular, FASN upregulation was identified as an early event during the development of prostate cancer (Swinnen et al., 2000b, 2002); evidence suggested that this lipogenic phenotype was driven by SREBF signaling (Swinnen et al., 2000a). Furthermore, RNAi silencing of FASN expression in an androgen-receptor-positive prostate cancer cell line strongly inhibited cell proliferation (De Schrijver et al., 2003). It has even been suggested that FASN itself can be sufficient to drive the transformation of prostate cells and may be a good target for antineoplastic therapy (Migita et al., 2009). Consequently, FASN has been the subject of drug development efforts, and specific FASN inhibitors, such as C75 and GSK837149A, have been developed and shown to kill cancer cells as well as synergize with established therapies (Menendez and Lupu, 2007). But crucially, these compounds have been shown to have poor pharmacokinetics or exhibit target-related toxicities. These findings support the hypothesis that targeting lipid synthesis can have a marked effect on cancer growth, but that the current selected targets may not be optimal, highlighting the need for the discovery of additional therapeutic targets that inhibit lipid metabolism. The sole carbon source and precursor for both fatty acid and cholesterol biosynthesis in mammalian cells is acetyl-CoA. The cytosolic pool of acetyl-CoA is mainly supplied by two different ATP-dependent reactions: cleavage of citrate into oxaloacetate and acetyl-CoA by ATP citrate lyase (ACLY) or the ligation of acetate and CoA by acetyl-CoA synthetase (ACSS). It has been shown that ACLY is required for cell growth and cancer cell survival, and there has been interest in the development of ACLY inhibitors, with some showing potential at inhibiting tumor growth (Beckers et al., 2007; Hatzivassiliou et al., 2005; Zu et al., 2012). However, only a few studies have addressed the potential role of acetate in the context of cancer (Yoshii et al., 2009a, 2009b; Yoshimoto et al., 2001; Yun et al., 2009). There are three genes that encode ACSS proteins, namely ACSS1, ACSS2, and a more recently proposed third family member ACSS3 (Luong et al., 2000; Watkins et al., 2007). ACSSs were originally identified as SREBF target genes, but relatively little is known regarding the regulation of ACSS gene expression (Luong et al., 2000; Schwer et al., 2006; Sone et al., 2002). Early studies indicated that ACSS activity controlled acetate uptake (Luong et al., 2000; Tucek, 1967). Herein, we attempted to identify lipid metabolism genes that were critical to the growth and survival of breast and prostate cancer cells in metabolically stressed conditions, such as hypoxia and low lipid availability. Results Exposure to Hypoxia and Low Serum Alters Lipid Metabolism It is now widely accepted that de novo lipogenesis is critical to tumor progression and survival and is driven by the increased expression of a host of lipogenic enzymes (reviewed by Menendez and Lupu, 2007; Zaidi et al., 2013). We initially characterized the dependency of breast cancer cell lines on de novo fatty acid synthesis under high- and low-serum conditions. The FASN inhibitor C75 had no effect on breast cancer cell lines when cultured in the presence of 10% serum (Figure 1A). However, treatment in low serum reduced cell number in the majority of the breast cancer cell lines tested. In addition, the IC50 for a recently developed FASN inhibitor, AZ22, was drastically improved when cells were cultured in low serum (Figure 1B and Figure S1A available online). To delineate changes in the lipid profiles of normoxic versus hypoxic cancer cells, we performed high-resolution acyl chain profiling of phosphatidylcholine (PC) and phosphatidylethanolamine (PE) using liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based lipidomics. We noted a shift in the profile of PC and PE that was mainly characterized by the presence of shorter, more saturated acyl chains in hypoxia compared to normoxia (Figures 1C and S1B). This type of shift is indicative of increased de novo fatty acid synthesis and elongation, which produce mainly saturated fatty acyl-CoA and, to a lesser degree, monounsaturated acyl-CoA (Hilvo et al., 2011; Rysman et al., 2010; Sahar et al., 2014). Since glucose can contribute to the synthesis of phospholipids by supplying the glycerol backbone via glycerol 3-phosphate and also by the production of the fatty acid precursor acetyl-CoA, we traced the fate of 13C6-glucose using targeted metabolomics and lipidomics (Figure S1C). It should be noted that all metabolomics experiments throughout the entire study used custom-formulated serum-like tissue culture medium (SMEM) containing concentrations of metabolites more physiologically relevant to human plasma than what is typically found in standard growth mediums. Selected metabolite concentrations were based on the most current literature where available and also as defined by the NIH (http://www.nlm.nih.gov/medlineplus/ency/article/003361.htm; Table S1). A high volume of medium to cell number ratio was maintained to ensure cells were exposed to concentrations of metabolites that were still within the reported physiological ranges. For instance, we found that 20% of available glucose in normoxia and 40% during hypoxia was consumed during a typical 24 hr incubation (Figure S1D). We also noted that, in normoxia, glucose was readily used by the tricarboxylic acid (TCA) cycle with >75% of citrate labeled from glucose; however, under hypoxic conditions, this contribution was decreased 5-fold (Figure S1E). This was likely due to the well-characterized inactivation of pyruvate dehydrogenase by pyruvate dehydrogenase kinase 1 under hypoxic conditions (Kim et al., 2006; Papandreou et al., 2006). In addition, an in-depth analysis of the most abundant PC species (34:1) detected a high degree of labeling under normoxic conditions (>95%), suggesting that the majority of PC(34:1) had been synthesized de novo from glucose (Figure S1F). Strikingly, in hypoxia, there was a sharp decline in the labeling of the fatty acid component of PC(34:1) (i.e., isotopologs ≥ M+4), but not the labeling of the glycerol backbone (M+3) (Figure S1F). These results imply that the majority of glycerol within the PC fraction is generated de novo by the cells, and suggest a strong dependency on biosynthetic pathways (Brockmöller et al., 2012). Given the observed changes in lipid metabolism and increased sensitivity to FASN inhibition in hypoxia and low serum, we next examined whether spheroids and tumor xenografts, models that naturally generate oxygen and nutrient gradients, also were susceptible to FASN inhibition. DU145, MDA-MB-468, and PC3 spheroid cultures treated with AZ22 were all dose-dependently sensitive to FASN inhibition (Figure 1D). Furthermore, using a different FASN inhibitor (AZ62) with improved pharmacokinetics and increased tolerance in vivo, we found that AZ62 inhibited BT474 xenograft tumor growth (Figures 1E and S1G). Analysis of metabolite extracts from the AZ62-treated tumors by liquid chromatography-mass spectrometry (LC-MS) detected elevated levels of the fatty acid precursor acetyl-CoA and the PC precursor CDP-choline (Figure 1F). Altogether, these results indicate that metabolic stress induces major changes in the lipid metabolism landscape of cancer cells, creating a dependence on de novo lipogenesis for survival in hypoxia/low serum (Ackerman and Simon, 2014; Bensaad et al., 2014). The data imply that, despite a high level of de novo lipogenesis under hypoxia (30%–50% labeled from glucose via glycerol 3-phosphate), glucose contribution to fatty acid biosynthesis is markedly decreased and an alternative carbon source would be required. Lastly, the growth inhibition observed in conditions of metabolic stress was recapitulated in spheroids and tumors, suggesting that tumors are indeed subject to conditions in which oxygen and lipids are severely limited. Functional Genomics Identifies ACSS2 as a Critical Gene for Cancer Cell Survival under Hypoxic or Low-Serum Conditions Expression of many genes involved in lipid biosynthesis is induced in response to activation of SREBF by the PI3K/Akt/mTORC1-signaling axis (Porstmann et al., 2008), which is frequently deregulated in human cancer. Furthermore, 50% of genes predicted to be highly significant for increasing the biomass of cancer cells by a genome-scale metabolic model of cancer were associated with lipid metabolism (Folger et al., 2011). Based on these independent lines of evidence, we assembled a small interfering RNA (siRNA) library of 66 potential targets with gene ontologies associated with lipid metabolism (Figure 2A; Tables S2 and S3). Functional genomics screens were performed using eight breast and three prostate cancer cell lines (Table S4). Based on our preliminary results with FASN inhibitors (Figure 1), we investigated growth inhibition during metabolic stress: 1% serum in normoxia, 10% serum in hypoxia, and 1% serum in hypoxia compared to the nutrient- and oxygen-rich conditions of 10% serum in normoxia. A strictly standardized mean difference (SSMD) was used to evaluate the potency and selectivity of silencing on growth relative to an internal nontargeting control (Zhang, 2011; Table S4). This preliminary filter was used to rank targets according to the difference in growth inhibition when comparing normoxia to hypoxia or 10% serum to 1% serum conditions (Table S4). The top targets from the screen were reassessed using a different pool of siRNAs (Figure 2B; Table S4). Following siRNA pool deconvolution of all the shortlisted targets, we found the top hits for hypoxia and low serum were, respectively, acetyl-CoA synthetase 2 (ACSS2) and stearoyl-CoA desaturase (SCD) (Figure 2B). We next performed a comprehensive data mining analysis of the expression and disease association of the targets that passed deconvolution. ACSS2 had the highest frequency of DNA copy-number gain in breast tumors and was associated with increased ACSS2 expression (Figures S2A and S2B). The relatively focal ACSS2 amplicon, which contains 47 other genes, spans most of the genomic region encompassing 20q11.22 and does not include any cancer consensus genes (Figures S2C and S2D). In support of the expression data from publically available patient data sets, immunohistochemical staining of tumor microarrays revealed high ACSS2 expression in nearly 40% of invasive ductal carcinomas (IDCs) compared to their normal adjacent tissue samples, with a similar trend observed in invasive lobular carcinomas (ILCs) (Figures 2C and 2D). Focused data mining of a separate breast cancer database noted a significant increase in ACSS2 copy number in cancer versus normal tissue (Figure 2E; Curtis et al., 2012). There was also a strong correlation between ACSS2 expression and disease progression (Figure 2F; Curtis et al., 2012). A Dunnet’s multiple comparison test between stage 0 and all other stages found there was significant upregulation of ACSS2 in stages 2, 3, and 4, and a test of linear trend further confirmed a systematic increase in ACSS2 expression with escalating tumor stage (p = 0.0001) (Figure 2F). Using this same data set, we further noted that ACSS2 expression was significantly higher (p < 0.05; Mann-Whitney test) in patients who did not survive past 1 or 5 years postdiagnosis (Figure 2G). In prostate cancer patients, there was an increased expression of ACSS2 in metastasis compared to primary tumor or normal tissue (Figures 2H and 2I). These results were consistent with observations from two different publically available prostate cancer data sets, suggesting that ACSS2 may be particularly important in more aggressive prostate cancers and metastases (Figure 2J). Hypoxia and Low-Serum Culture Conditions Synergistically Enhance ACSS2 Expression The Cancer Cell Line Encyclopedia was used to identify breast cancer cell lines with strong evidence for ACSS2 copy-number gain and upregulation (Barretina et al., 2012; Forbes et al., 2011; Shao et al., 2013). Two of the cancer cell lines used in the functional screen had high ACSS2 expression, namely MDA-MB-468 and SKBr3, while BT474 had the overall highest level of DNA copy-number gain and expression (Figures S3A and S3B). DNA copy-number gain for both the 20q arm and 20q11 chromosomal band, containing ACSS2, has been reported previously for BT474 and SKBr3, as detected by fluorescence in situ hybridization, genomic hybridization, and comparative genomic hybridization array (Brinkmann et al., 1996; Guan et al., 1996; Pinkel et al., 1998; Rondón-Lagos et al., 2014). Given the enhanced growth inhibition observed by ACSS2 silencing during the functional screening in hypoxia, we measured ACSS2-dependent growth during metabolic stress. Silencing of ACSS2 was most effective at growth inhibition under metabolic stress and, interestingly, was cytotoxic to BT474 cells (Figures 3A and S3C). Furthermore, expression analysis of our panel of cancer cell lines revealed consistent upregulation of ACSS2 at both the mRNA (Figures 3B, S3D, and S3E) and protein levels (Figure 3C) under metabolic stress. These results reinforced the hypothesis that ACSS2 becomes important for cell proliferation under metabolic stress and that cancer cells harboring ACSS2 copy-number gains may be more vulnerable to ACSS2 depletion. A test for mutual exclusivity among 962 breast cancer samples (The Cancer Genome Atlas) revealed a significant tendency toward co-occurrence between ACSS2 and hypoxia-inducible transcription factor (HIF) targets LDHA and PDK1, suggesting it may be a HIF target (Figure S3F). Indeed, small hairpin RNA (shRNA)-mediated silencing of ARNT expression reduced the upregulation of ACSS2 by 2.5-fold (Figures 3D and S3G). However, unlike the complete loss of hypoxia-induced upregulation of LDHA and PDK1 noted after ARNT silencing, some ACSS2 upregulation persisted (Figures 3D and S3H). Since ACSS2 is known to be a target of SREBF signaling (Luong et al., 2000) and SREBFs are activated by lipid depletion (Briggs et al., 1993; Wang et al., 1993), it seemed likely that ACSS2 expression would be controlled by SREBFs. Silencing of SREBF2, but not SREBF1, completely eliminated the upregulation of ACSS2 under conditions of metabolic stress (Figures 3E and S3I). Together, the expression data imply that SREBF2 mainly controls ACSS2 expression, but that ARNT-dependent HIF signaling enhances the upregulation of ACSS2 by SREBF2. Interestingly, the expression of FASN was highly upregulated under metabolic stress and controlled in exactly the same manner as ACSS2, suggesting the two targets are intimately linked (Figure S3J). Acetate Supports Biosynthesis of Membrane Phospholipids The molecular function of ACSS2 is to ligate acetate with coenzyme A in an ATP-dependent reaction to produce acetyl-CoA, a metabolite central to a range of cellular processes (Figure 4A). We investigated if acetate contributed to the acetyl-CoA pool and, furthermore, if it was used for lipid synthesis. To that end, BT474c1, a more tumorigenic subclone of BT474, was used. BT474c1 cells, which express similar levels of ACSS2 as their parental cells (Figure S3B), had increased 14C2-acetate uptake under hypoxic conditions (Figure 4B). Also, 14C2-acetate-dependent fatty acid synthesis increased during metabolic stress (Figure 4C). However, in all cell lines tested, the steady-state levels of acetyl-CoA were much lower in hypoxia (Figure 4D). To investigate this further, we traced the incorporation of 13C2-acetate into the acetyl-CoA pool and found that, at most, 10% was labeled during normoxic and lipid-replete growth conditions, while during metabolic stress, over 50% of the acetyl-CoA pool was labeled by acetate over a 12 hr period (Figure 4E). The mutual mRNA regulation of ACSS2 and FASN suggested acetate was being used to support membrane phospholipid biosynthesis during metabolic stress. To study this, we employed LC-MS/MS-based lipidomics to trace the incorporation of 13C2-acetate into the PC(34:1) during metabolic stress. There was a rightward shift of the PC-isotopolog-labeling pattern, indicating that a higher percentage of PC carbon was sourced from acetate during metabolic stress than normal growth conditions (Figure 4F). This shift is highly indicative of an increased availability of acetate-derived acetyl-CoA for fatty acid synthesis, and, importantly, shows that acetate can be a major nutritional source. ACSS2 Controls Acetate Uptake and Contribution to Fatty Acids Previous reports had suggested that acetate consumption is dictated by functional expression of ACSS2 (Luong et al., 2000; Tucek, 1967). Measurement of [3H]-acetate uptake in BT474c1 cells, as well as DU145 prostate cancer cells, demonstrated reduced acetate consumption after ACSS2 depletion (Figures 5A and 5B). Interestingly, BT474c1 cells that expressed high levels of ACSS2 exhibited almost twice as much acetate uptake compared to DU145 cells that expressed low levels of ACSS2 (Figures 5B and S3B). We further investigated for signs of inhibition of lipid synthesis by quantifying the substrate of the final enzymatic reaction in the synthesis of PC and PE (Figure 5C). Similar to the results using FASN inhibitors (Figure 1F), CDP-choline and CDP-ethanolamine steady-state levels were elevated upon ACSS2 silencing, suggesting impairment of de novo lipogenesis (Figure 5D). Since serum is a source of lipids, growth factors, and other nutrients, we reassessed ACSS2-dependent growth in lipid-depleted serum (LPDS) to ascertain if the observed growth effects during hypoxia, such as seen in Figure 3, were due to the loss of extracellular lipids or another component in the serum. Cells grown in 10% LPDS were more sensitive to ACSS2 silencing than cells grown in 10% full serum in hypoxia (Figure 5E). Similar to 1% serum, 10% LPDS also induced ACSS2 expression in hypoxia (Figure 5F). The most highly characterized pathway of acetate metabolism concerns the conversion of acetate to acetyl-CoA and the use of 2-carbon acetyl units to synthesize fatty acids through successive rounds of condensation by FASN (Roncari, 1974, 1975). We investigated if acetate was being used as an anabolic precursor for de novo fatty acid synthesis. The concentration of plasma acetate in humans varies between 0.05 and 0.25 mM (Scheppach et al., 1991; Skutches et al., 1979; Tollinger et al., 1979), but can increase up to 1 mM after alcohol consumption (Lundquist et al., 1962; Tsukamoto et al., 1989). The acetate concentration in mouse and rat plasma ranges between 0.20 and 0.30 mM (Kimura et al., 2013; Maxwell et al., 2010). The acetate concentration in media + 10% serum was measured by gas chromatography-mass spectrometry and determined to be 0.10 mM (Table S1). Besides exogenous sources, acetate can be produced endogenously via protein deacetylation. However, we and others observed that histone acetylation is induced under hypoxia indicating that deacetylation is unlikely an acetate source during metabolically stressed conditions (Vaapil et al., 2012; Watson et al., 2009; our unpublished observations). Initially, we characterized the contribution of 13C2-acetate to fatty acid synthesis by comparing the isotopolog-labeling pattern of palmitate and stearate during metabolic stress. LC-MS-based analysis of palmitate revealed small dose-dependent differences in acetate-dependent palmitate synthesis as the concentration of acetate increased from 0.10 to 0.50 mM (Figure 5G). Similar to the PC-isotopolog-labeling pattern in Figure 4, there was an overall shift in the abundance of isotopologs < M+8 to ≥ M+8 when cells were exposed to metabolic stress, suggesting increased availability of acetate-derived acetyl-CoA for fatty acid synthesis (Figure 5G). Similar results were obtained for stearate (Figure S4A). In addition, silencing of ACSS2 strongly impaired both palmitate and stearate labeling from acetate (Figures S4B and S4C). We next traced the fate of 13C2-acetate within the TCA cycle by looking at the isotpolog-labeling pattern of citrate. However, this would be controlled mainly by the other ACSS isoform, ACSS1, which has been shown to be localized to the mitochondria (Fujino et al., 2001). ACSS1 activity did slightly compensate for silencing of ACSS2 under normoxic conditions, in which an active TCA cycle was still running (Figure S4D). This was shown by the increase in M+2 and M+3 labeling of citrate following ACSS2 silencing in normoxia (Figure S4D). However, under hypoxic conditions, the compensation by ACSS1 was compromised (Figure S4D). This is in line with the observation that ACSS1 mRNA expression was downregulated during hypoxia, which is completely opposite to the mRNA regulation of ACSS2 (Figure S4E). Subsequently, we supplied cells with 13C6-glucose, 13C5-glutamine, or 13C2-acetate to compare the relative contributions of these important nutritional sources to palmitate synthesis. In normal growth conditions, glucose was the main source for fatty acid synthesis, followed closely by acetate (Figure 5H). In contrast, metabolic stress switched the preferred nutritional source to acetate and completely abrogated the contribution from glucose (Figure 5H). As previously reported, there was an increase in glutamine-labeled palmitate during hypoxic conditions (Figure 5H; Filipp et al., 2012; Metallo et al., 2012; Mullen et al., 2012; Wise et al., 2011). However, neither glucose nor glutamine, whether in normal growth conditions or low oxygen and low serum, ever labeled palmitate as extensively as acetate did during metabolic stress. ACSS2 Silencing Inhibits Spheroid and Tumor Growth DU145 + shNTC and DU145 + shACSS2#64 cells were cultured as spheroids in 10% or 1% serum and in the presence or absence of doxycycline. Expression of shACSS2 by doxycycline decreased ACSS2 expression and caused significant growth inhibition, which was more pronounced under low-serum conditions (Figure 6A). We had observed previously that acetate was being used to generate lipids (Figures 4 and 5). In addition, the FASN inhibitor studies and spheroid growth after ACSS2 silencing prompted us to investigate the in vivo efficacy of ACSS2 silencing on tumor growth and survival. For this we also generated a BT474c1 cell line expressing inducible shRNA targeting ACSS2 (Figure 5A). Nude mice were injected subcutaneously with DU145, MDA-MB-468, and BT474c1 cells expressing ACSS2 shRNA or a nontargeting control or empty vector. All three xenograft models displayed decreases in tumor growth when ACSS2 was silenced (Figures 6B, 6C, S5A, and S5B). Gene silencing was confirmed by immunohistochemistry or quantitative RT-PCR (qRT-PCR) (Figures 6D, S5C, and S5D). We also noted that, by the end of the xenograft studies, all mice bearing tumors depleted of ACSS2 (i.e., +DOX) were still viable, while nontreated or vector control mice exhibited higher mortality rates, which were not necessarily associated with tumor burden (Figure 6E). Our previous results suggested that ACSS2 expression was often upregulated under hypoxic conditions (Figure 3). To examine this in vivo, serial sections from three separate BT474c1 + shACSS2#20 (no DOX) tumors were stained with pimonidazole, a hypoxia-selective marker, and ACSS2. Indeed, costaining was observed, indicating that hypoxic tumor regions have significantly higher levels of ACSS2 compared to nonhypoxic regions (Figures 6F and 6G). Together, these results confirm that ACSS2 is upregulated in areas of hypoxia in tumors and required for efficient growth and survival of cancer cells in vivo. Discussion Hypoxia is a common feature of tumors that have a poor clinical prognosis and display aggressive traits, such as increased metastatic and migratory potential, chemoresistance, and stemness. We also know that lipid metabolism is altered drastically under these conditions (Ackerman and Simon, 2014). Our initial studies identified that most breast and prostate cancer cell lines were sensitive to fatty acid synthesis inhibition when cultured as spheroids or grown as tumor xenografts. However, the only tissue culture condition that faithfully preserved this sensitivity was when cells were grown under metabolic stress, thus, strongly suggesting that many cancer cells find a means to survive and proliferate regardless of poor nutrient delivery and hypoxia. The functional screens and in-vivo-induced silencing of ACSS2 in tumor xenografts highlighted the essentiality of ACSS2 under metabolically stressed conditions. The metabolic and lipidomic analyses revealed a mechanistic solution for the need of acetate and ACSS2 in cancer cells by demonstrating that, under tumor-like tissue culture conditions, there is a switch in nutrient utilization from glucose to acetate to support fatty acid and lipid biosynthesis. In support of this mechanism, we found that ACSS2 was indeed significantly upregulated in hypoxic regions of tumor xenografts and when cancer cells were exposed to prolonged oxygen and lipid withdrawal. ACSS2 expression was transcriptionally controlled by a synergistic interaction between HIF and SREBF2 signaling. In addition, copy-number gain of ACSS2 was observed in breast carcinoma, and the lack of cancer consensus genes and the size of the narrow amplicon region suggested selection pressure. The data mining information was supported by results from examining tissue microarrays showing that ACSS2 was highly expressed in IDCs, the most common histological breast cancer subtype. Likewise, increased ACSS2 expression seemed to be more closely associated with prostate metastasis than in primary tumors. Our observed link between acetate metabolism and tumor growth coupled with the knowledge that the primary source of acetate production within the body is by the intestinal microbiota compelled us to investigate if there was a potential link between colorectal cancers and ACSS2. Further cancer data mining revealed copy-number gain or upregulation of ACSS2 in more than a quarter of all colorectal cancer (data not shown). If ACSS2 silencing proves to be effective at inhibiting colorectal tumor growth, it may reveal a disease in which ACSS2 would be an attractive target for future drug discovery. Furthermore, in cells with active lipid biosynthesis, acetate is converted to acetyl-CoA in the cytosol and then incorporated into fatty acids. Therefore, radiolabeled acetate is the most lipid-specific tracer available for positron emission tomography (PET) imaging. Acetate-based PET tracers are established clinical tools, but to date there is not enough information on how this tracer reflects perturbations in lipid biosynthesis in cancer cells (Lewis et al., 2014; Oyama et al., 2002; Yun et al., 2009). We have identified a dependency of breast cancers and prostate cancer on acetate metabolism and ACSS2 activity. This presents an attractive opportunity to use tracer technology to identify tumors that would be responsive to ACSS2-targeted therapy. Moreover, as acetate uptake is coupled to ACSS2 activity, acetate PET tracers could provide an important pharmacodynamic biomarker for treatment response and for detecting when tumors become refractory to treatment. In conclusion, this study highlights the importance of understanding the context-dependent metabolic rewiring of cancer cells in vivo, and provides evidence that acetate is a fundamental nutrient that can fuel cancer growth. The therapeutic feasibility of pharmacologically targeting ACSS2 is currently being explored. Experimental Procedures Cell Culture LNCaP were obtained from the American Type Culture Collection. BT474c1 cells were a kind gift from Jose Baselga. All other cell lines were from the London Research Institute (CRUK) cell services. DU145, LNCaP, and PC3 cell lines were grown in RPMI supplemented with either 10% or 1% fetal calf serum (PAA Laboratories), 2 mM L-glutamine, and penicillin/streptomycin. MCF7, BT20, BT474, BT474c1, BT549, MDA-MB-231, MDA-MB-468, SkBr3, and T47D cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM)/F12 with either 10% or 1% fetal calf serum (PAA Laboratories), 2 mM L-glutamine, and penicillin/streptomycin. MCF10a and MCF10A + HER2 (a kind gift from Dr. Zachary Schafer) cells were grown in DMEM/F12 with either 5% or 0.5% horse serum with 20 ng/ml epidermal growth factor, 5 μg/ml hydrocortisone, 10 μg/ml insulin, and 100 ng/ml cholera toxin. Antibodies and Reagents ACSS2 (AceS1) antibody (D19C6) and FASN were purchased from Cell Signaling Technology. Antitubulin, antiactin, anti-ACSS1 (HPA043228), and anti-ACSS2 (HPA004141) antibodies were purchased from Sigma-Aldrich. HypoxyProbe kit, including pimonidazole, was purchased from Chemicon International. All chemicals were purchased from Fisher Scientific or Sigma-Aldrich. The 13C-labeled metabolites were purchased from Cambridge Isotopes, and 14C-labeled and 3H-labeled acetate were purchased from PerkinElmer. Secondary antibodies were purchased from LI-COR Biosciences. Spheroid Growth Assay For spheroid formation, cells were trypsinized, counted, mixed with 2% matrigel in culture medium, and placed in 96-well ultralow attachment plates (Costar). Spheroid formation was initiated by centrifugation at 850 × g for 10 min, and cultures were incubated for the times indicated. Fresh growth medium, containing either ethanol or doxycycline (0.5 mg/ml), was administered every 48 hr. Spheroid size was determined at the times indicated by automated imaging on an inverted microscope (Axiovert 100M, Carl Zeiss). Xenograft Experiments Female BALB/c nude mice (Charles River Laboratories) were injected subcutaneously with BT474, BT474c1, DU145, or MDA-MB-468 cells and allowed to establish for 1 week. When BT474 or BT474c1 cells were used, mice were implanted with 17-beta-estradiol 0.36 mg/pellet 24 hr prior to injection of cells. For shRNA induction, animals were given a doxycycline-containing diet (Harlan Laboratories) or doxycycline-supplemented drinking water, and tumor growth was followed for the indicated time period. Tumor volume was determined by caliper measurements and the ellipsoidal volume formula: ½ × length × width2. Where available, tumor burden also was determined using bioluminescence imaging at indicated times. Mice were maintained in individually ventilated cages with environmental enrichment. All procedures were performed with anesthetic and postoperative analgesics under the Animal (Scientific Procedures) Act 1986 and the EU Directive 2010. Animal experiments at Imperial College London were carried out under UK Home Office Project License 70/7177 (E.O.A.) with ethical approval from Imperial College London within a Designated Establishment under the Act (the Central Biomedical Services Unit at Imperial College London). Animal experiments at the London Research Institute were performed under UK Home Office Project License 80/2330 (A.S.) after approval by a local ethics committee. Animal experiments at the Beatson Institute were performed under UK Home Office Project License 60/4181 (K.B.) and carried out with ethical approval from University of Glasgow. LC-MS Cells were plated onto 12-well plates and cultured in standard medium DMEM/F12 + 10% fetal bovine serum and 2 mM glutamine. Medium was replaced for specified periods with fresh physiologically relevant medium containing amino acid concentrations as dictated by http://www.nlm.nih.gov/medlineplus/ency/article/003361.htm. In addition, all heavy carbon labeling was done using final concentrations of 5.5 mM glucose, or 0.65 mM glutamine, and 0.10–0.50 mM acetate. Metabolites were extracted with a cold solution (−20°C) composed of methanol, acetonitrile, and water (5:3:2). The insoluble material was immediately pelleted in a cooled (0°C) centrifuge at 16,000 × g for 15 min, and the supernatant was collected for subsequent LC-MS analysis. The protein concentration in each well was calculated using a Lowry assay. Alternatively, a separate plate was seeded in parallel and used for cell counts. A ZIC-pHILIC column (4.6 × 150 mm, guard column 2.1 × 20 mm, Merck) was used for LC separation using formic acid, water, and acetonitrile as component of the mobile phase. Metabolites were detected using Thermo Exactive mass spectrometer (Thermo Fisher Scientific). For lipidomic studies, cells were incubated in media containing 5.5 mM 13C6-glucose for 24 hr prior to lipid isolation. Cell pellets were washed twice with cold degassed PBS and resuspended in methanol before transfer to silanized glass tubes on dry ice to Babraham Lipidomics Facility for lipid extraction and analysis. Samples were spiked with 400 ng 12:0/12:0-PC, 100 ng 17:0-LPC, 300 ng 12:0/12:0-PE, and 100 ng 17:1-LPE; 0.88% NaCl; and chloroform. The mixture was vortexed for 20 s at room temperature and sonicated in an ice-cold water bath for 2 min. Samples were centrifuged at 1,100 rpm at 4°C for 15 min. The lower phase was collected. The upper phase was extracted with synthetic lower phase (mix chloroform/methanol/0.88% NaCl at volume ratio of 2:1:1, after phase separation, take the lower phase as synthetic lower phase for second extraction of lipid). The resulting lower phase was combined and dried under vacuum at room temperature with SpeedVac (Thermo Scientific) and redissolved in chloroform. The final product was injected for LC-MS/MS analysis using a Thermo Orbitrap Elite system (Thermo Fisher Scientific) hyphenated with a five-channel online degasser, four-pump, column oven, and autosampler with cooler Prominence HPLC system (Shimadzu) for lipids analysis. In detail, lipid classes were separated on a normal-phase silica gel column (2.1 × 150 mm, 4micro, MicoSolv Technology) with hexane/dichloromethane/chloroform/methanol/acetanitrile/water/ethylamine solvent gradient based on the polarity of head group. High resolution (240k at m/z 400)/accurate mass (with mass accuracy < 5 ppm) and tandem MS (collision-induced fragmentation) were used for molecular species identification and quantification. The identity of lipid was further confirmed by reference to appropriate lipids standards. See also the Supplemental Experimental Procedures. Author Contributions Z.T.S and B.P. performed the growth assays, interpreted the results, and wrote the manuscript. Z.T.S, B.P., and S.G. performed data mining. D.T.J. performed all spheroid experiments and and I.S.A. contributed acetate uptake and AZ62 in vivo studies. Z.T.S., Q.Z., and E.S. performed lipidomics. Z.T.S., N.J.F.v.d.B., and G.M. performed metabolomics. Z.T.S., V.B., N.J.F.v.d.B., G.M., and J.J.K. peformed GC/MS. Z.T.S performed immunohistochemistry of tumor sections and 14C2-acetate experiments. Z.T.S., B.P., and D.T.J. performed western blots. Z.T.S. and B.P. performed qRT-PCR analyses. Z.T.S., B.P., L.M., D.J., E.S., M.J., and M.H. performed and analyzed the functional genomic screens. G.K. and R.E.S. calculated SSMDs. B.P., B.S.-D., and G.S. analyzed the tissue microarrays. Z.T.S, S.M., K.B., F.L., and M.Z.T. did the bioluminescent imaging. Z.T.S. and D.S. quantified the ACSS2 and pimonidazole costaining. S.T. formulated SMEM. L.M.G. performed the pIC50 experiment. A.L.H., E.O.A., S.E.C., M.J.O.W., A.S., E.G., Z.T.S, B.P., D.T.J., and Q.Z. contributed to the interpretation and conceptual advancement through quarterly face-to-face meetings.