- Record: found
- Abstract: found
- Article: found
PNPLA3 variant and portal/periportal histological pattern in patients with biopsy-proven non-alcoholic fatty liver disease: a possible role for oxidative stress
Read this article at
Abstract
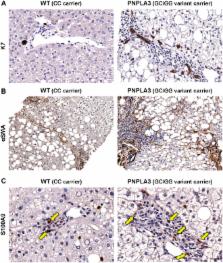
Pathogenesis of non-alcoholic fatty liver disease (NAFLD) is influenced by predisposing genetic variations, dysmetabolism, systemic oxidative stress, and local cellular and molecular cross-talks. Patatin-like phospholipase domain containing 3 (PNPLA3) gene I148M variant is a known determinant of NAFLD. Aims were to evaluate whether PNPLA3 I148M variant was associated with a specific histological pattern, hepatic stem/progenitor cell (HpSC) niche activation and serum oxidative stress markers. Liver biopsies were obtained from 54 NAFLD patients. The activation of HpSC compartment was evaluated by the extension of ductular reaction (DR); hepatic stellate cells, myofibroblasts (MFs), and macrophages were evaluated by immunohistochemistry. Systemic oxidative stress was assessed measuring serum levels of soluble NOX2-derived peptide (sNOX2-dp) and 8-isoprostaglandin F 2α (8-iso-PGF 2α). PNPLA3 carriers showed higher steatosis, portal inflammation and HpSC niche activation compared to wild-type patients. DR was correlated with NAFLD activity score (NAS) and fibrosis score. Serum 8-iso-PGF 2α were significantly higher in I148M carriers compared to non-carriers and were correlated with DR and portal inflammation. sNox2-dp was correlated with NAS and with HpSC niche activation. In conclusion, NAFLD patients carrying PNPLA3 I148M are characterized by a prominent activation of HpSC niche which is associated with a more aggressive histological pattern (portal fibrogenesis) and increased oxidative stress.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
Liver regeneration - mechanisms and models to clinical application.
- Record: found
- Abstract: found
- Article: not found