- Record: found
- Abstract: found
- Article: found
Cell-Associated Flagella Enhance the Protection Conferred by Mucosally-Administered Attenuated Salmonella Paratyphi A Vaccines
Read this article at
Abstract
Background
Antibiotic-resistant Salmonella enterica serovar Paratyphi A, the agent of paratyphoid A fever, poses an emerging public health dilemma in endemic areas of Asia and among travelers, as there is no licensed vaccine. Integral to our efforts to develop a S. Paratyphi A vaccine, we addressed the role of flagella as a potential protective antigen by comparing cell-associated flagella with exported flagellin subunits expressed by attenuated strains.
Methodology
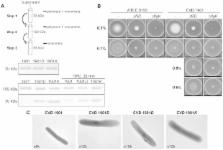
S. Paratyphi A strain ATCC 9150 was first deleted for the chromosomal guaBA locus, creating CVD 1901. Further chromosomal deletions in fliD (CVD 1901D) or flgK (CVD 1901K) were then engineered, resulting in the export of unpolymerized FliC, without impairing its overall expression. The virulence of the resulting isogenic strains was examined using a novel mouse LD 50 model to accommodate the human-host restricted S. Paratyphi A. The immunogenicity of the attenuated strains was then tested using a mouse intranasal model, followed by intraperitoneal challenge with wildtype ATCC 9150.
Results
Mucosal (intranasal) immunization of mice with strain CVD 1901 expressing cell-associated flagella conferred superior protection (vaccine efficacy [VE], 90%) against a lethal intraperitoneal challenge, compared with the flagellin monomer-exporting mutants CVD 1901K (30% VE) or CVD 1901D (47% VE). The superior protection induced by CVD 1901 with its cell-attached flagella was associated with an increased IgG2a∶IgG1 ratio of FliC-specific antibodies with enhanced opsonophagocytic capacity.
Conclusions
Our results clearly suggest that enhanced anti-FliC antibody-mediated clearance of S. Paratyphi A by phagocytic cells, induced by vaccines expressing cell-associated rather than exported FliC, might be contributing to the vaccine-induced protection from S. Paratyphi A challenge in vivo. We speculate that an excess of IgG1 anti-FliC antibodies induced by the exported FliC may compete with the IgG2a subtype and block binding to specific phagocyte Fc receptors that are critical for clearing an S. Paratyphi A infection.
Author Summary
Salmonella enterica serovar Paratyphi A is a pathogen that causes a systemic disease that is marked by serious complications and, if untreated, high mortality. The study of S. Paratyphi A pathogenesis and vaccine development has been extremely challenging since S. Paratyphi A is human host-restricted and no appropriate animal model exists. Since there is currently no licensed vaccine to prevent paratyphoid fever caused by this organism, our study represents a pioneering attempt to develop and refine a vaccine against S. Paratyphi A. We employed live attenuated strains which allow in vivo presentation of bacterial antigens via the natural route of infection, without the complications associated with antigen production and purification for subunit vaccines. For determining protective immunity against infection, we developed a mouse model that allowed evaluation of vaccine efficacy. We used our system to examine the protective capacity of a major Salmonella antigen, the flagellum. Due to its unique immunogenic properties, the flagellum is considered a major immune mediator, but its role in protection is controversial. We clearly show that cell-associated flagellar protein, presented by mucosally administered attenuated bacterial live vaccines, provides superior protection when compared to strains exporting FliC monomers, and we discuss possible mechanisms of immunity.
Related collections
Most cited references52
- Record: found
- Abstract: found
- Article: not found
Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf.
- Record: found
- Abstract: found
- Article: not found
Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid.
- Record: found
- Abstract: found
- Article: not found