- Record: found
- Abstract: found
- Article: found
A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting
Read this article at
Abstract
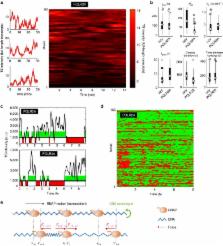
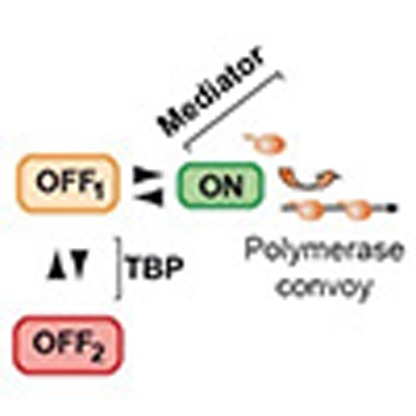
Live-cell imaging has revealed unexpected features of gene expression. Here using improved single-molecule RNA microscopy, we show that synthesis of HIV-1 RNA is achieved by groups of closely spaced polymerases, termed convoys, as opposed to single isolated enzymes. Convoys arise by a Mediator-dependent reinitiation mechanism, which generates a transient but rapid succession of polymerases initiating and escaping the promoter. During elongation, polymerases are spaced by few hundred nucleotides, and physical modelling suggests that DNA torsional stress may maintain polymerase spacing. We additionally observe that the HIV-1 promoter displays stochastic fluctuations on two time scales, which we refer to as multi-scale bursting. Each time scale is regulated independently: Mediator controls minute-scale fluctuation (convoys), while TBP-TATA-box interaction controls sub-hour fluctuations (long permissive/non-permissive periods). A cellular promoter also produces polymerase convoys and displays multi-scale bursting. We propose that slow, TBP-dependent fluctuations are important for phenotypic variability of single cells.
Abstract
 HIV-1 viral gene expression stochastically switches between active and inactive states.
Here, using improved single molecule RNA microscopy, the authors show that HIV-1 RNA
stochastic transcription is achieved by groups of closely spaced polymerases, and
is regulated by Mediator and TBP at different time scales.
HIV-1 viral gene expression stochastically switches between active and inactive states.
Here, using improved single molecule RNA microscopy, the authors show that HIV-1 RNA
stochastic transcription is achieved by groups of closely spaced polymerases, and
is regulated by Mediator and TBP at different time scales.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
Nature, nurture, or chance: stochastic gene expression and its consequences.
- Record: found
- Abstract: found
- Article: not found
Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise.

- Record: found
- Abstract: found
- Article: found