- Record: found
- Abstract: found
- Article: found
Recent advances in dynamic m 6A RNA modification
Read this article at
Abstract
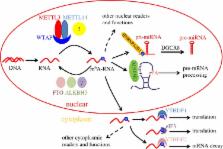
The identification of m 6A demethylases and high-throughput sequencing analysis of methylated transcriptome corroborated m 6A RNA epigenetic modification as a dynamic regulation process, and reignited its investigation in the past few years. Many basic concepts of cytogenetics have been revolutionized by the growing understanding of the fundamental role of m 6A in RNA splicing, degradation and translation. In this review, we summarize typical features of methylated transcriptome in mammals, and highlight the ‘writers’, ‘erasers’ and ‘readers’ of m 6A RNA modification. Moreover, we emphasize recent advances of biological functions of m 6A and conceive the possible roles of m 6A in the regulation of immune response and related diseases.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase.
- Record: found
- Abstract: found
- Article: not found
Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells.
- Record: found
- Abstract: found
- Article: not found