- Record: found
- Abstract: found
- Article: found
CRISPR/Cas9 nickase-mediated disruption of hepatitis B virus open reading frame S and X
Read this article at
Abstract
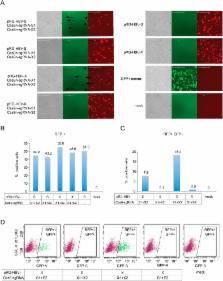
Current antiviral therapies cannot cure hepatitis B virus (HBV) infection; successful HBV eradication would require inactivation of the viral genome, which primarily persists in host cells as episomal covalently closed circular DNA (cccDNA) and, to a lesser extent, as chromosomally integrated sequences. However, novel designer enzymes, such as the CRISPR/Cas9 RNA-guided nuclease system, provide technologies for developing advanced therapy strategies that could directly attack the HBV genome. For therapeutic application in humans, such designer nucleases should recognize various HBV genotypes and cause minimal off-target effects. Here, we identified cross-genotype conserved HBV sequences in the S and X region of the HBV genome that were targeted for specific and effective cleavage by a Cas9 nickase. This approach disrupted not only episomal cccDNA and chromosomally integrated HBV target sites in reporter cell lines, but also HBV replication in chronically and de novo infected hepatoma cell lines. Our data demonstrate the feasibility of using the CRISPR/Cas9 nickase system for novel therapy strategies aiming to cure HBV infection.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
Hepatitis B virus infection.
- Record: found
- Abstract: found
- Article: not found
Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects.
- Record: found
- Abstract: found
- Article: not found