- Record: found
- Abstract: found
- Article: not found
Modeling Biphasic Environmental Decay of Pathogens and Implications for Risk Analysis

Read this article at
Abstract
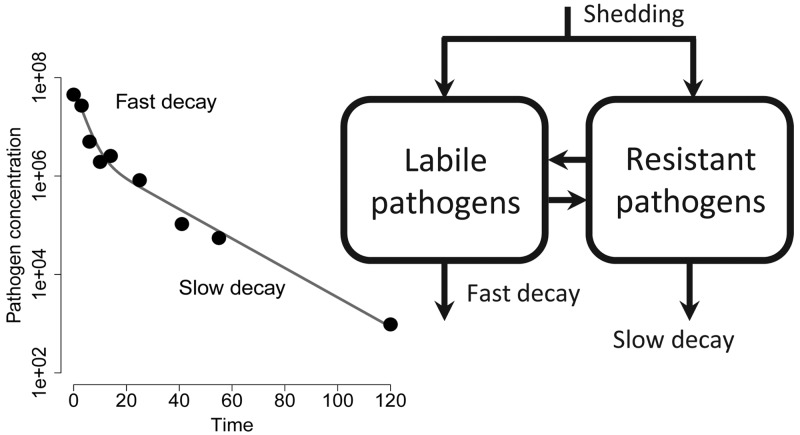
As the appreciation for the importance of the environment in infectious disease transmission has grown, so too has interest in pathogen fate and transport. Fate has been traditionally described by simple exponential decay, but there is increasing recognition that some pathogens demonstrate a biphasic pattern of decay—fast followed by slow. While many have attributed this behavior to population heterogeneity, we demonstrate that biphasic dynamics can arise through a number of plausible mechanisms. We examine the identifiability of a general model encompassing three such mechanisms: population heterogeneity, hardening off, and the existence of viable-but-not-culturable states. Although the models are not fully identifiable from longitudinal sampling studies of pathogen concentrations, we use a differential algebra approach to determine identifiable parameter combinations. Through case studies using Cryptosporidium and Escherichia coli, we show that failure to consider biphasic pathogen dynamics can lead to substantial under- or overestimation of disease risks and pathogen concentrations, depending on the context. More reliable models for environmental hazards and human health risks are possible with an improved understanding of the conditions in which biphasic die-off is expected. Understanding the mechanisms of pathogen decay will ultimately enhance our control efforts to mitigate exposure to environmental contamination.
Related collections
Most cited references60
- Record: found
- Abstract: found
- Article: not found
Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood.
- Record: found
- Abstract: found
- Article: not found
Dormancy contributes to the maintenance of microbial diversity.
- Record: found
- Abstract: not found
- Article: not found
