- Record: found
- Abstract: found
- Article: not found
Molecular evolution of the ent-kaurenoic acid oxidase gene in Oryzeae
Read this article at
Abstract
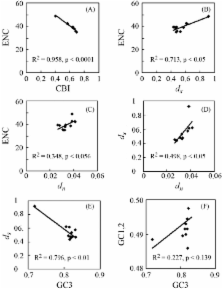
We surveyed the substitution patterns in the ent-kaurenoic acid oxidase (KAO) gene in 11 species of Oryzeae with an outgroup in the Ehrhartoidaea. The synonymous and non-synonymous substitution rates showed a high positive correlation with each other, but were negatively correlated with codon usage bias and GC content at third codon positions. The substitution rate was heterogenous among lineages. Likelihood-ratio tests showed that the non-synonymous/synonymous rate ratio changed significantly among lineages. Site-specific models provided no evidence for positive selection of particular amino acid sites in any codon of the KAO gene. This finding suggested that the significant rate heterogeneity among some lineages may have been caused by variability in the relaxation of the selective constraint among lineages or by neutral processes.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
The origins of genome complexity.
- Record: found
- Abstract: found
- Article: not found
The 'effective number of codons' used in a gene.
- Record: found
- Abstract: found
- Article: not found
