- Record: found
- Abstract: found
- Article: found
RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors
Read this article at
Abstract
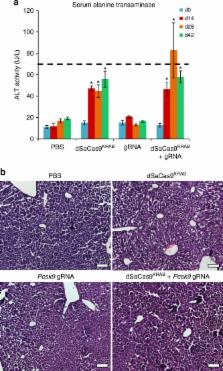
CRISPR-Cas9 transcriptional repressors have emerged as robust tools for disrupting gene regulation in vitro but have not yet been adapted for systemic delivery in adult animal models. Here we describe a Staphylococcus aureus Cas9-based repressor (dSaCas9 KRAB) compatible with adeno-associated viral (AAV) delivery. To evaluate dSaCas9 KRAB efficacy for gene silencing in vivo, we silenced transcription of Pcsk9, a regulator of cholesterol levels, in the liver of adult mice. Systemic administration of a dual-vector AAV8 system expressing dSaCas9 KRAB and a Pcsk9-targeting guide RNA (gRNA) results in significant reductions of serum Pcsk9 and cholesterol levels. Despite a moderate host response to dSaCas9 KRAB expression, Pcsk9 repression is maintained for 24 weeks after a single treatment, demonstrating the potential for long-term gene silencing in post-mitotic tissues with dSaCas9 KRAB. In vivo programmable gene silencing enables studies that link gene regulation to complex phenotypes and expands the CRISPR-Cas9 perturbation toolbox for basic research and gene therapy applications.
Abstract
Repression of gene transcription using CRISPR-Cas9 has been achieved in vitro but not for delivery into adult animal models. Here, the authors use AAV8 to deliver the transcriptional repressor dSaCas9 KRAB to the cholesterol regulator Pcsk9, and show repression up to 24 weeks and reduced cholesterol levels in mice.
Related collections
Most cited references16
- Record: found
- Abstract: found
- Article: not found
CRISPR RNA-guided activation of endogenous human genes
- Record: found
- Abstract: found
- Article: not found
In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9.
- Record: found
- Abstract: found
- Article: not found