- Record: found
- Abstract: found
- Article: found
Modulation of transcriptional burst frequency by histone acetylation
Read this article at
Significance
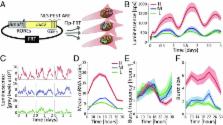
Single-cell approaches have shown that many mammalian genes are transcribed stochastically in bursts of specific sizes and frequencies; however, molecular mechanisms controlling these bursting parameters have remained largely undetermined. By studying transcriptional bursting of a luciferase reporter controlled by a circadian gene promoter, we found that the gene integration site mainly influenced the burst size, while the circadian time primarily modulated the burst frequency. These daily variations in burst frequency correlated with histone acetylation levels, and CRISPR-Cas9–mediated acetylation of the promoter was sufficient to change the burst frequency. Since this correlation was also observed in other genes and in several cell types, we conclude that the impact of histone acetylation on gene expression is achieved mainly through modulation of burst frequency.
Abstract
Many mammalian genes are transcribed during short bursts of variable frequencies and sizes that substantially contribute to cell-to-cell variability. However, which molecular mechanisms determine bursting properties remains unclear. To probe putative mechanisms, we combined temporal analysis of transcription along the circadian cycle with multiple genomic reporter integrations, using both short-lived luciferase live microscopy and single-molecule RNA-FISH. Using the Bmal1 circadian promoter as our model, we observed that rhythmic transcription resulted predominantly from variations in burst frequency, while the genomic position changed the burst size. Thus, burst frequency and size independently modulated Bmal1 transcription. We then found that promoter histone-acetylation level covaried with burst frequency, being greatest at peak expression and lowest at trough expression, while remaining unaffected by the genomic location. In addition, specific deletions of ROR-responsive elements led to constitutively elevated histone acetylation and burst frequency. We then investigated the suggested link between histone acetylation and burst frequency by dCas9p300-targeted modulation of histone acetylation, revealing that acetylation levels influence burst frequency more than burst size. The correlation between acetylation levels at the promoter and burst frequency was also observed in endogenous circadian genes and in embryonic stem cell fate genes. Thus, our data suggest that histone acetylation-mediated control of transcription burst frequency is a common mechanism to control mammalian gene expression.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Nature, nurture, or chance: stochastic gene expression and its consequences.
- Record: found
- Abstract: found
- Article: not found
Epigenome editing by a CRISPR/Cas9-based acetyltransferase activates genes from promoters and enhancers
- Record: found
- Abstract: found
- Article: not found