- Record: found
- Abstract: found
- Article: found
Using drug descriptions and molecular structures for drug–drug interaction extraction from literature
Read this article at
Abstract
Motivation
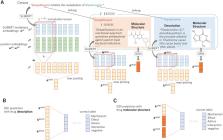
Neural methods to extract drug–drug interactions (DDIs) from literature require a large number of annotations. In this study, we propose a novel method to effectively utilize external drug database information as well as information from large-scale plain text for DDI extraction. Specifically, we focus on drug description and molecular structure information as the drug database information.
Results
We evaluated our approach on the DDIExtraction 2013 shared task dataset. We obtained the following results. First, large-scale raw text information can greatly improve the performance of extracting DDIs when combined with the existing model and it shows the state-of-the-art performance. Second, each of drug description and molecular structure information is helpful to further improve the DDI performance for some specific DDI types. Finally, the simultaneous use of the drug description and molecular structure information can significantly improve the performance on all the DDI types. We showed that the plain text, the drug description information and molecular structure information are complementary and their effective combination is essential for the improvement.
Related collections
Most cited references24

- Record: found
- Abstract: found
- Article: found
DrugBank 5.0: a major update to the DrugBank database for 2018
- Record: found
- Abstract: not found
- Article: not found
Scikit-learn: Machine Learning in Python
- Record: found
- Abstract: not found
- Article: not found