- Record: found
- Abstract: found
- Article: found
The Atlantic salmon ( Salmo salar) antimicrobial peptide cathelicidin-2 is a molecular host-associated cue for the salmon louse ( Lepeophtheirus salmonis)
Read this article at
Abstract
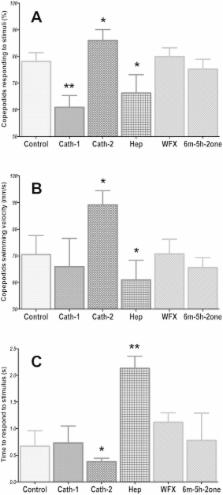
Chemical signals are a key element of host-parasite interactions. In marine ecosystems, obligate ectoparasites, such as sea lice, use chemical cues and other sensory signals to increase the probability of encountering a host and to identify appropriate hosts on which they depend to complete their life cycle. The chemical compounds that underlie host identification by the sea lice are not fully described or characterized. Here, we report a novel compound - the Atlantic salmon ( Salmo salar) antimicrobial peptide cathelicidin-2 (Cath-2) – that acts as an activation cue for the marine parasitic copepod Lepeophtheirus salmonis. L. salmonis were exposed to 0, 7, 70 and 700 ppb of Cath-2 and neural activity, swimming behaviour and gene expression profiles of animals in response to the peptide were evaluated. The neurophysiological, behavioural and transcriptomic results were consistent: L. salmonis detects Cath-2 as a water-soluble peptide released from the skin of salmon, triggering chemosensory neural activity associated with altered swimming behaviour of copepodids exposed to the peptide, and chemosensory-related genes were up-regulated in copepodids exposed to the peptide. L. salmonis are activated by Cath-2, indicating a tight link between this peptide and the salmon louse chemosensory system.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Sensing odorants and pheromones with chemosensory receptors.
- Record: found
- Abstract: found
- Article: not found
Ecology of sea lice parasitic on farmed and wild fish.
- Record: found
- Abstract: not found
- Article: not found