- Record: found
- Abstract: found
- Article: found
The Sequence Alignment/Map format and SAMtools
Read this article at
Abstract
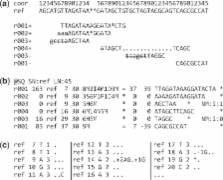
Summary: The Sequence Alignment/Map (SAM) format is a generic alignment format for storing read alignments against reference sequences, supporting short and long reads (up to 128 Mbp) produced by different sequencing platforms. It is flexible in style, compact in size, efficient in random access and is the format in which alignments from the 1000 Genomes Project are released. SAMtools implements various utilities for post-processing alignments in the SAM format, such as indexing, variant caller and alignment viewer, and thus provides universal tools for processing read alignments.
Availability: http://samtools.sourceforge.net
Contact: rd@ 123456sanger.ac.uk