- Record: found
- Abstract: found
- Article: not found
Chlamydia preserves the mitochondrial network necessary for replication via microRNA-dependent inhibition of fission
Read this article at
Abstract
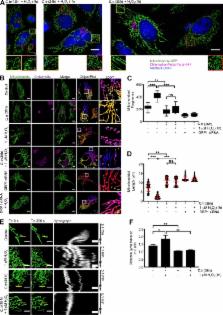
Chlamydiae are intracellular pathogens that depend on the host for their survival and development. Chowdhury et al. demonstrate that Chlamydia trachomatis infection can prevent mitochondrial fission in primary cells by reducing DRP1 abundance via miR-30c–dependent inhibition of p53.
Abstract
Obligate intracellular bacteria such as Chlamydia trachomatis depend on metabolites of the host cell and thus protect their sole replication niche by interfering with the host cells’ stress response. Here, we investigated the involvement of host microRNAs (miRNAs) in maintaining the viability of C. trachomatis–infected primary human cells. We identified miR-30c-5p as a prominently up-regulated miRNA required for the stable down-regulation of p53, a major suppressor of metabolite supply in C. trachomatis–infected cells. Loss of miR-30c-5p led to the up-regulation of Drp1, a mitochondrial fission regulator and a target gene of p53, which, in turn, severely affected chlamydial growth and had a marked effect on the mitochondrial network. Drp1-induced mitochondrial fragmentation prevented replication of C. trachomatis even in p53-deficient cells. Additionally, Chlamydia maintain mitochondrial integrity during reactive oxygen species–induced stress that occurs naturally during infection. We show that C. trachomatis require mitochondrial ATP for normal development and hence postulate that they preserve mitochondrial integrity through a miR-30c-5p–dependent inhibition of Drp1-mediated mitochondrial fission.
Related collections
Most cited references68
- Record: found
- Abstract: found
- Article: not found
Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis.
- Record: found
- Abstract: found
- Article: not found
Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells.

- Record: found
- Abstract: found
- Article: found
