- Record: found
- Abstract: found
- Article: found
Genome of a Novel Bacterium “ Candidatus Jettenia ecosi” Reconstructed From the Metagenome of an Anammox Bioreactor
Read this article at
Abstract
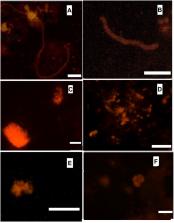
The microbial community of a laboratory-scale bioreactor based on the anammox process was investigated by using metagenomic approaches and fluorescent in situ hybridization (FISH). The bioreactor was initially inoculated with activated sludge from the denitrifying bioreactor of a municipal wastewater treatment station. By constantly increasing the ammonium and nitrite load, a microbial community containing the novel species of anammox bacteria “ Candidatus Jettenia ecosi” developed in the bioreactor after 5 years when the maximal daily nitrogen removal rate reached 8.5 g/L. Sequencing of the metagenome of anammox granules and the binning of the contigs obtained, allowed a high quality draft genome of the dominant anammox bacterium, “ Candidatus Jettenia ecosi” to be assembled. Annotation of the 3.9 Mbp long genome revealed 3970 putative protein-coding genes, 45 tRNA genes, and genes for 16S/23S rRNAs. Analysis of the genome of “ Candidatus Jettenia ecosi” revealed genes involved in anammox metabolism, including nitrite and ammonium transporters, copper-containing nitrite reductase, a nitrate reductase complex, hydrazine synthase, and hydrazine dehydrogenase. Autotrophic carbon fixation could be accomplished through the Wood Ljungdahl pathway. The composition of the community was investigated through a search of 16S rRNA sequences in the metagenome and FISH analysis of the anammox granules. The presence of the members of Ignavibacteriae, Betaproteobacteria, Chloroflexi and other microbial lineages reflected the complexity of the microbial processes in the studied bioreactor performed by anammox Planctomycetes, fermentative bacteria, and denitrifiers.
Related collections
Most cited references57

- Record: found
- Abstract: found
- Article: found
Ribosomal Database Project: data and tools for high throughput rRNA analysis
- Record: found
- Abstract: found
- Article: not found