- Record: found
- Abstract: found
- Article: found
Cleavage mediated by the P15 domain of bacterial RNase P RNA
Read this article at
Abstract
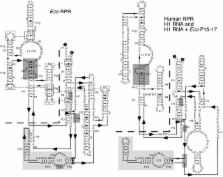
Independently folded domains in RNAs frequently adopt identical tertiary structures regardless of whether they are in isolation or are part of larger RNA molecules. This is exemplified by the P15 domain in the RNA subunit (RPR) of the universally conserved endoribonuclease P, which is involved in the processing of tRNA precursors. One of its domains, encompassing the P15 loop, binds to the 3′-end of tRNA precursors resulting in the formation of the RCCA–RNase P RNA interaction (interacting residues underlined) in the bacterial RPR–substrate complex. The function of this interaction was hypothesized to anchor the substrate, expose the cleavage site and result in re-coordination of Mg 2+ at the cleavage site. Here we show that small model-RNA molecules (~30 nt) carrying the P15-loop mediated cleavage at the canonical RNase P cleavage site with significantly reduced rates compared to cleavage with full-size RPR. These data provide further experimental evidence for our model that the P15 domain contributes to both substrate binding and catalysis. Our data raises intriguing evolutionary possibilities for ‘RNA-mediated’ cleavage of RNA.
Related collections
Most cited references59
- Record: found
- Abstract: found
- Article: not found
Tertiary contacts distant from the active site prime a ribozyme for catalysis.
- Record: found
- Abstract: found
- Article: not found
Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA
- Record: found
- Abstract: found
- Article: not found