- Record: found
- Abstract: found
- Article: found
Ancient origin of a Western Mediterranean radiation of subterranean beetles
Read this article at
Abstract
Background
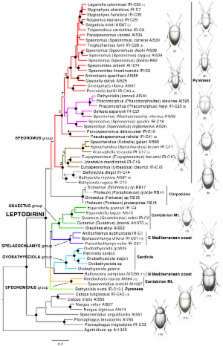
Cave organisms have been used as models for evolution and biogeography, as their reduced above-ground dispersal produces phylogenetic patterns of area distribution that largely match the geological history of mountain ranges and cave habitats. Most current hypotheses assume that subterranean lineages arose recently from surface dwelling, dispersive close relatives, but for terrestrial organisms there is scant phylogenetic evidence to support this view. We study here with molecular methods the evolutionary history of a highly diverse assemblage of subterranean beetles in the tribe Leptodirini (Coleoptera, Leiodidae, Cholevinae) in the mountain systems of the Western Mediterranean.
Results
Ca. 3.5 KB of sequence information from five mitochondrial and two nuclear gene fragments was obtained for 57 species of Leptodirini and eight outgroups. Phylogenetic analysis was robust to changes in alignment and reconstruction method and revealed strongly supported clades, each of them restricted to a major mountain system in the Iberian peninsula. A molecular clock calibration of the tree using the separation of the Sardinian microplate (at 33 MY) established a rate of 2.0% divergence per MY for five mitochondrial genes (4% for cox1 alone) and dated the nodes separating the main subterranean lineages before the Early Oligocene. The colonisation of the Pyrenean chain, by a lineage not closely related to those found elsewhere in the Iberian peninsula, began soon after the subterranean habitat became available in the Early Oligocene, and progressed from the periphery to the centre.
Conclusions
Our results suggest that by the Early-Mid Oligocene the main lineages of Western Mediterranean Leptodirini had developed all modifications to the subterranean life and were already present in the main geographical areas in which they are found today. The origin of the currently recognised genera can be dated to the Late Oligocene-Miocene, and their diversification can thus be traced to Miocene ancestors fully adapted to subterranean life, with no evidence of extinct epigean, less modified lineages. The close correspondence of organismal evolution and geological record confirms them as an important study system for historical biogeography and molecular evolution.
Related collections
Most cited references35
- Record: found
- Abstract: found
- Article: not found
An algorithm for progressive multiple alignment of sequences with insertions.
- Record: found
- Abstract: not found
- Article: not found
Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times.
- Record: found
- Abstract: found
- Article: not found