- Record: found
- Abstract: found
- Article: found
Description of a new species of Aplectana (Nematoda: Ascaridomorpha: Cosmocercidae) using an integrative approach and preliminary phylogenetic study of Cosmocercidae and related taxa

Read this article at
Abstract
Background
Nematodes of the family Cosmocercidae (Ascaridomorpha: Cosmocercoidea) are mainly parasitic in the digestive tract of various amphibians and reptiles worldwide. However, our knowledge of the molecular phylogeny of the Cosmocercidae is still far from comprehensive. The phylogenetic relationships between Cosmocercidae and the other two families, Atractidae and Kathlaniidae, in the superfamily Cosmocercoidea are still under debate. Moreover, the systematic position of some genera within Cosmocercidae remains unclear.
Methods
Nematodes collected from Polypedates megacephalus (Hallowell) (Anura: Rhacophoridae) were identified using morphological (light and scanning electron microscopy) and molecular methods [sequencing the small ribosomal DNA (18S), internal transcribed spacer 1 (ITS-1), large ribosomal DNA (28S) and mitochondrial cytochrome c oxidase subunit 1 ( cox1) target regions]. Phylogenetic analyses of cosmocercoid nematodes using 18S + 28S sequence data were performed to clarify the phylogenetic relationships of the Cosmocercidae, Atractidae and Kathlaniidae in the Cosmocercoidea and the systematic position of the genus Aplectana in Cosmocercidae.
Results
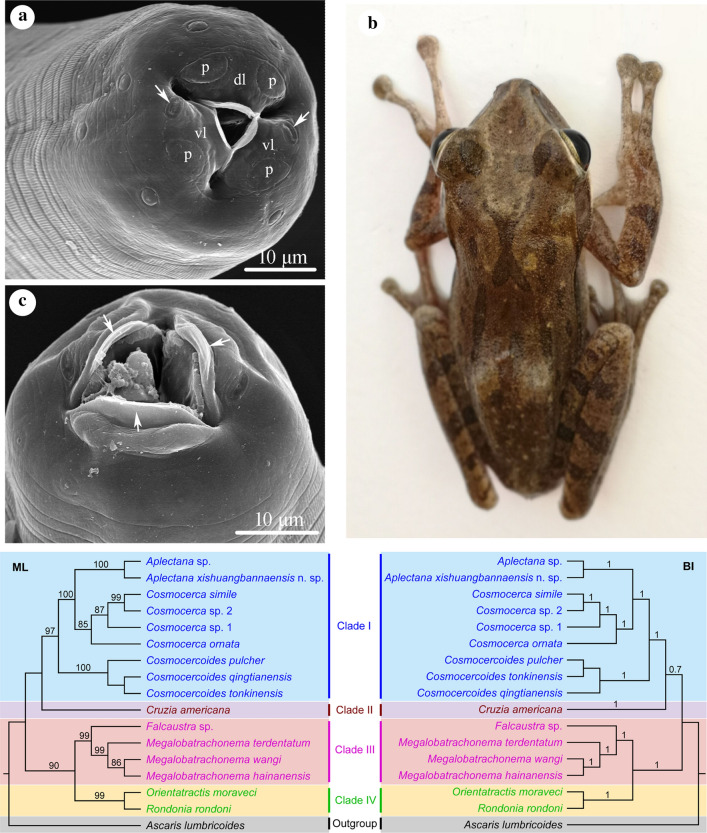
Morphological and genetic evidence supported the hypothesis that the nematode specimens collected from P. megacephalus represent a new species of Aplectana (Cosmocercoidea: Cosmocercidae). Our phylogenetic results revealed that the Cosmocercidae is a monophyletic group, but not the basal group in Cosmocercoidea as in the traditional classification. The Kathlaniidae is a paraphyletic group because the subfamily Cruziinae within Kathlaniidae (including only the genus Cruzia) formed a seperate lineage. Phylogenetic analyses also showed that the genus Aplectana has a closer relationship to the genus Cosmocerca in Cosmocercidae.
Conclusions
Our phylogenetic results suggested that the subfamily Cruziinae should be moved from the hitherto-defined family Kathlaniidae and elevated as a separate family, and the genus Cosmocerca is closely related to the genus Aplectana in the family Cosmocercidae. The present study provided a basic molecular phylogenetic framework for the superfamily Cosmocercoidea based on 18S + 28S sequence data for the first time to our knowledge. Moreover, a new species, A. xishuangbannaensis n. sp., was described using integrative approach.
Related collections
Most cited references41

- Record: found
- Abstract: found
- Article: found
IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies
- Record: found
- Abstract: found
- Article: found
MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space
- Record: found
- Abstract: not found
- Article: not found