- Record: found
- Abstract: found
- Article: found
CRL4–DCAF1 ubiquitin E3 ligase directs protein phosphatase 2A degradation to control oocyte meiotic maturation
Read this article at
Abstract
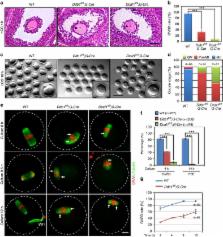
Oocyte meiosis is a specialized cell cycle that gives rise to fertilizable haploid gametes and is precisely controlled in various dimensions. We recently found that E3 ubiquitin ligase CRL4 is required for female fertility by regulating DNA hydroxymethylation to maintain oocyte survival and to promote zygotic genome reprogramming. However, not all phenotypes of CRL4-deleted oocytes could be explained by this mechanism. Here we show that CRL4 controls oocyte meiotic maturation by proteasomal degradation of protein phosphatase 2A scaffold subunit, PP2A-A. Oocyte-specific deletion of DDB1 or DCAF1 (also called VPRBP) results in delayed meiotic resumption and failure to complete meiosis I along with PP2A-A accumulation. DCAF1 directly binds to and results in the poly-ubiquitination of PP2A-A. Moreover, combined deletion of Ppp2r1a rescues the meiotic defects caused by DDB1/DCAF1 deficiency. These results provide in vivo evidence that CRL4-directed PP2A-A degradation is physiologically essential for regulating oocyte meiosis and female fertility.
Abstract
 The E3 ubiquitin ligase CRL4 regulates oocyte survival through hydroxymethylation
of genomic DNA. Here Yu
et al. show that CRL4 is also required for oocytes to complete meiosis I by mediating the
poly-ubiquitination and proteasomal degradation of the cell cycle regulator protein
phosphatase 2A-A subunit.
The E3 ubiquitin ligase CRL4 regulates oocyte survival through hydroxymethylation
of genomic DNA. Here Yu
et al. show that CRL4 is also required for oocytes to complete meiosis I by mediating the
poly-ubiquitination and proteasomal degradation of the cell cycle regulator protein
phosphatase 2A-A subunit.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase.
- Record: found
- Abstract: found
- Article: not found
Global identification of modular cullin-RING ligase substrates.
- Record: found
- Abstract: found
- Article: not found