- Record: found
- Abstract: found
- Article: found
Pressure-Overload Cardiac Hypertrophy Is Associated with Distinct Alternative Splicing Due to Altered Expression of Splicing Factors
Read this article at
Abstract
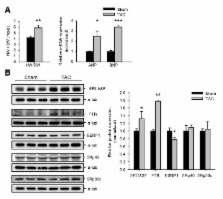
Chronic pressure-overload cardiac hypertrophy is associated with an increased risk of morbidity/mortality, largely due to maladaptive remodeling and dilatation that progresses to dilated cardiomyopathy. Alternative splicing is an important biological mechanism that generates proteomic complexity and diversity. The recent development of next-generation RNA sequencing has improved our understanding of the qualitative signatures associated with alternative splicing in various biological conditions. However, the role of alternative splicing in cardiac hypertrophy is yet unknown. The present study employed RNA-Seq and a bioinformatic approach to detect the RNA splicing regulatory elements involved in alternative splicing during pressure-overload cardiac hypertrophy. We found GC-rich exonic motifs that regulate intron retention in 5′ UTRs and AT-rich exonic motifs that are involved in exclusion of the AT-rich elements that cause mRNA instability in 3′ UTRs. We also identified motifs in the intronic regions involved in exon exclusion and inclusion, which predicted splicing factors that bind to these motifs. We found, through Western blotting, that the expression levels of three splicing factors, ESRP1, PTB and SF2/ASF, were significantly altered during cardiac hypertrophy. Collectively, the present results suggest that chronic pressure-overload hypertrophy is closely associated with distinct alternative splicing due to altered expression of splicing factors.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Predictive identification of exonic splicing enhancers in human genes.
- Record: found
- Abstract: found
- Article: not found
A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition.
- Record: found
- Abstract: found
- Article: not found