- Record: found
- Abstract: found
- Article: found
Asymptomatic Intestinal Colonization with Protist Blastocystis Is Strongly Associated with Distinct Microbiome Ecological Patterns
Read this article at
Abstract
Given the results of our study and other reports of the effects of the most common human gut protist on the diversity and composition of the bacterial microbiome, Blastocystis and, possibly, other gut protists should be studied as ecosystem engineers that drive community diversity and composition.
ABSTRACT
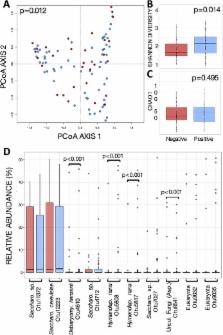
Blastocystis is the most prevalent protist of the human intestine, colonizing approximately 20% of the North American population and up to 100% in some nonindustrialized settings. Blastocystis is associated with gastrointestinal and systemic disease but can also be an asymptomatic colonizer in large populations. While recent findings in humans have shown bacterial microbiota changes associated with this protist, it is unknown whether these occur due to the presence of Blastocystis or as a result of inflammation. To explore this, we evaluated the fecal bacterial and eukaryotic microbiota in 156 asymptomatic adult subjects from a rural population in Xoxocotla, Mexico. Colonization with Blastocystis was strongly associated with an increase in bacterial alpha diversity and broad changes in beta diversity and with more discrete changes to the microbial eukaryome. More than 230 operational taxonomic units (OTUs), including those of dominant species Prevotella copri and Ruminococcus bromii, were differentially abundant in Blastocystis-colonized individuals. Large functional changes accompanied these observations, with differential abundances of 202 (out of 266) predicted metabolic pathways (PICRUSt), as well as lower fecal concentrations of acetate, butyrate, and propionate in colonized individuals. Fecal calprotectin was markedly decreased in association with Blastocystis colonization, suggesting that this ecological shift induces subclinical immune consequences to the asymptomatic host. This work is the first to show a direct association between the presence of Blastocystis and shifts in the gut bacterial and eukaryotic microbiome in the absence of gastrointestinal disease or inflammation. These results prompt further investigation of the role Blastocystis and other eukaryotes play within the human microbiome.
IMPORTANCE Given the results of our study and other reports of the effects of the most common human gut protist on the diversity and composition of the bacterial microbiome, Blastocystis and, possibly, other gut protists should be studied as ecosystem engineers that drive community diversity and composition.
Related collections
Most cited references57

- Record: found
- Abstract: found
- Article: found
Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data
- Record: found
- Abstract: found
- Article: not found
New insights on classification, identification, and clinical relevance of Blastocystis spp.
- Record: found
- Abstract: found
- Article: not found