- Record: found
- Abstract: found
- Article: not found
High-Throughput In Vivo Analysis of Gene Expression in Caenorhabditis elegans
Read this article at
Abstract
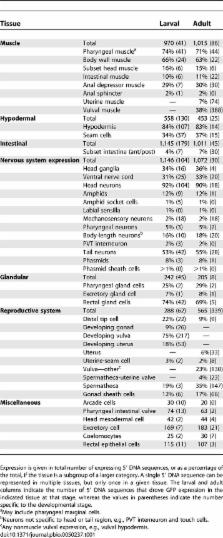
Using DNA sequences 5′ to open reading frames, we have constructed green fluorescent protein (GFP) fusions and generated spatial and temporal tissue expression profiles for 1,886 specific genes in the nematode Caenorhabditis elegans. This effort encompasses about 10% of all genes identified in this organism. GFP-expressing wild-type animals were analyzed at each stage of development from embryo to adult. We have identified 5′ DNA regions regulating expression at all developmental stages and in 38 different cell and tissue types in this organism. Among the regulatory regions identified are sequences that regulate expression in all cells, in specific tissues, in combinations of tissues, and in single cells. Most of the genes we have examined in C. elegans have human orthologs. All the images and expression pattern data generated by this project are available at WormAtlas ( http://gfpweb.aecom.yu.edu/index) and through WormBase ( http://www.wormbase.org).
Author Summary
Knowing where a protein is expressed provides an important clue about its potential function. As critical as this information is, we have complete developmental expression profiles for only a small fraction of all genes expressed in any metazoan. Here, we have generated spatial and temporal tissue expression profiles for 10% of all genes in the nematode Caenorhabditis elegans. Worms expressing putative gene regulatory elements fused with green fluorescent protein were analyzed at each stage of development from embryo to adult. Among the regulatory regions identified are sequences that regulate expression in all cells, in specific tissues, in combinations of tissues, and in single cells. Most of the genes we have examined in C. elegans have human orthologs. Our analysis of complex expression patterns for so many genes may not only facilitate functional analysis in C. elegans, but also create a foundation for decoding the informational hierarchies governing gene expression in all organisms.
Abstract
Using DNA sequences 5' to open reading frames, the authors construct green fluorescent protein fusions and generate spatial and temporal tissue expression profiles for 10% of all genes in the nematode Caenorhabditis elegans.
Related collections
Most cited references43
- Record: found
- Abstract: found
- Article: not found
Genome sequence of the nematode C. elegans: a platform for investigating biology.
- Record: found
- Abstract: found
- Article: not found
A gene expression atlas of the central nervous system based on bacterial artificial chromosomes.
- Record: found
- Abstract: found
- Article: not found
