- Record: found
- Abstract: found
- Article: not found
The High Mobility Group (Hmg) Boxes of the Nuclear Protein Hmg1 Induce Chemotaxis and Cytoskeleton Reorganization in Rat Smooth Muscle Cells
Read this article at
Abstract
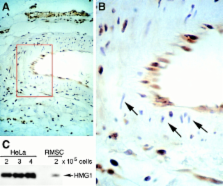
HMG1 (high mobility group 1) is a ubiquitous and abundant chromatin component. However, HMG1 can be secreted by activated macrophages and monocytes, and can act as a mediator of inflammation and endotoxic lethality. Here we document a role of extracellular HMG1 in cell migration. HMG1 (and its individual DNA-binding domains) stimulated migration of rat smooth muscle cells in chemotaxis, chemokinesis, and wound healing assays. HMG1 induced rapid and transient changes of cell shape, and actin cytoskeleton reorganization leading to an elongated polarized morphology typical of motile cells. These effects were inhibited by antibodies directed against the receptor of advanced glycation endproducts, indicating that the receptor of advanced glycation endproducts is the receptor mediating the HMG1-dependent migratory responses. Pertussis toxin and the mitogen-activated protein kinase kinase inhibitor PD98059 also blocked HMG1-induced rat smooth muscle cell migration, suggesting that a G i/o protein and mitogen-activated protein kinases are required for the HMG1 signaling pathway. We also show that HMG1 can be released by damage or necrosis of a variety of cell types, including endothelial cells. Thus, HMG1 has all the hallmarks of a molecule that can promote atherosclerosis and restenosis after vascular damage.
Related collections
Most cited references43
- Record: found
- Abstract: found
- Article: not found
HMG-1 as a late mediator of endotoxin lethality in mice.
- Record: found
- Abstract: found
- Article: not found
Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa.
- Record: found
- Abstract: found
- Article: not found
